Project
DANIO-CODE
Navigation
Downloads
Results for arid5b
Z-value: 1.11
Transcription factors associated with arid5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arid5b
|
ENSDARG00000037196 | AT-rich interaction domain 5B |
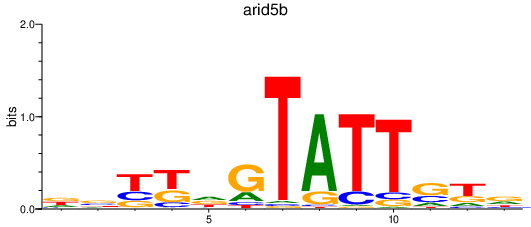
Activity profile of arid5b motif
Sorted Z-values of arid5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arid5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_2296253 | 3.13 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr22_+_818795 | 3.13 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr18_-_16964811 | 2.60 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr14_-_8634381 | 2.32 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr7_-_73613531 | 2.29 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr2_-_57339717 | 2.21 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr5_-_14000166 | 1.95 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr19_-_10411573 | 1.92 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr13_-_6123969 | 1.92 |
ENSDART00000161163
|
tuba4l
|
tubulin, alpha 4 like |
| chr14_+_8634518 | 1.90 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr3_-_40142513 | 1.79 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr15_+_44168257 | 1.78 |
ENSDART00000176254
|
CU655961.7
|
ENSDARG00000106724 |
| chr25_-_17822003 | 1.74 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr13_+_46652067 | 1.72 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr9_+_38715293 | 1.70 |
ENSDART00000131846
|
osbpl11
|
oxysterol binding protein-like 11 |
| chr18_-_321203 | 1.58 |
ENSDART00000166810
|
tm2d3
|
TM2 domain containing 3 |
| chr25_+_20596490 | 1.58 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr25_-_24960874 | 1.45 |
ENSDART00000178891
|
ENSDARG00000061421
|
ENSDARG00000061421 |
| chr14_-_20940726 | 1.43 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr15_-_37687982 | 1.39 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr2_-_44924505 | 1.36 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr25_-_17822194 | 1.33 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr18_+_36801847 | 1.31 |
ENSDART00000004129
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr12_+_31668168 | 1.26 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr9_+_2019992 | 1.26 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr7_+_13238684 | 1.25 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr5_-_71605608 | 1.24 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr14_+_50010976 | 1.23 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr16_+_29574449 | 1.23 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr4_+_18525469 | 1.20 |
ENSDART00000154154
|
BX649398.1
|
ENSDARG00000097195 |
| chr10_-_244746 | 1.19 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr18_+_18417187 | 1.17 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr22_-_31089831 | 1.16 |
|
|
|
| chr21_+_6890129 | 1.15 |
|
|
|
| chr19_-_10411518 | 1.13 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr15_-_37687790 | 1.12 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr24_-_36248243 | 1.10 |
ENSDART00000065336
|
tmem14cb
|
transmembrane protein 14Cb |
| chr24_-_28358514 | 1.10 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr2_-_37762426 | 1.04 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr21_-_41508736 | 1.03 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr12_+_34790513 | 1.03 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr21_-_30217360 | 1.01 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr4_+_90885 | 0.99 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr7_-_37283894 | 0.99 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr4_-_765957 | 0.98 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr14_-_16118942 | 0.97 |
|
|
|
| chr15_-_37687938 | 0.95 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr4_+_30530 | 0.95 |
ENSDART00000157825
|
syn3
|
synapsin III |
| chr9_+_21448920 | 0.94 |
ENSDART00000137024
|
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr5_+_63596241 | 0.92 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr6_-_46733412 | 0.92 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr9_-_30174641 | 0.86 |
ENSDART00000134157
ENSDART00000089206 |
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr11_+_7173167 | 0.83 |
ENSDART00000125619
|
thop1
|
thimet oligopeptidase 1 |
| chr5_+_71100066 | 0.83 |
ENSDART00000115182
|
nup214
|
nucleoporin 214 |
| chr16_+_27289472 | 0.82 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr19_+_19806658 | 0.81 |
ENSDART00000160444
|
si:dkey-95j14.1
|
si:dkey-95j14.1 |
| chr5_-_68751287 | 0.77 |
ENSDART00000112692
|
ENSDARG00000077155
|
ENSDARG00000077155 |
| chr15_-_5636514 | 0.76 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr9_+_48535267 | 0.75 |
ENSDART00000141405
|
fastkd1
|
FAST kinase domains 1 |
| chr5_+_13370060 | 0.75 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr7_+_22521055 | 0.74 |
ENSDART00000146801
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr7_+_22520928 | 0.74 |
ENSDART00000146801
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr14_+_15778201 | 0.73 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr7_+_26730804 | 0.73 |
|
|
|
| chr20_-_51829168 | 0.73 |
ENSDART00000177319
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr9_+_14052296 | 0.71 |
ENSDART00000124267
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr18_-_7488960 | 0.70 |
ENSDART00000052803
|
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr17_+_14956899 | 0.70 |
|
|
|
| chr2_+_31823314 | 0.70 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr23_-_27663219 | 0.69 |
ENSDART00000138381
|
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr16_-_22397159 | 0.66 |
|
|
|
| chr19_-_19806070 | 0.65 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr15_+_44168674 | 0.63 |
|
|
|
| chr21_+_7543440 | 0.62 |
|
|
|
| chr19_-_10411878 | 0.61 |
ENSDART00000136653
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr7_+_73595871 | 0.60 |
ENSDART00000169756
|
HIST1H2BA (1 of many)
|
Histone H2B 1/2 |
| chr15_-_34810397 | 0.60 |
|
|
|
| chr3_-_40142333 | 0.59 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr25_-_2263905 | 0.58 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr15_-_34810486 | 0.58 |
|
|
|
| chr14_+_8634323 | 0.57 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr18_+_7584201 | 0.57 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr23_+_36608246 | 0.55 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr19_-_12058542 | 0.55 |
ENSDART00000168308
|
si:ch73-49k18.1
|
si:ch73-49k18.1 |
| chr8_+_36467811 | 0.54 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr16_+_27289443 | 0.53 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr9_-_2886135 | 0.53 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr10_-_35331891 | 0.50 |
|
|
|
| chr21_+_43177310 | 0.49 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr5_+_63596451 | 0.49 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr10_+_38981942 | 0.49 |
|
|
|
| chr7_+_17464608 | 0.48 |
ENSDART00000173792
|
rtn3
|
reticulon 3 |
| chr13_-_30020072 | 0.46 |
ENSDART00000143093
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr14_-_38441710 | 0.45 |
|
|
|
| chr18_-_20159345 | 0.44 |
|
|
|
| chr2_-_50391964 | 0.43 |
ENSDART00000009347
|
exoc3
|
exocyst complex component 3 |
| chr5_-_23727236 | 0.43 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr15_+_1018191 | 0.43 |
|
|
|
| chr5_+_58327436 | 0.43 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr9_-_35073289 | 0.43 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr2_-_6545431 | 0.42 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr2_-_37702092 | 0.42 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr5_-_71605690 | 0.41 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr12_-_25006119 | 0.39 |
ENSDART00000158036
|
cript
|
cysteine-rich PDZ-binding protein |
| chr21_+_1084935 | 0.36 |
|
|
|
| chr8_+_22323986 | 0.34 |
ENSDART00000062254
|
MAD2L2
|
mitotic arrest deficient 2 like 2 |
| chr5_+_13370237 | 0.33 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr23_-_29579049 | 0.33 |
ENSDART00000165744
|
kif1b
|
kinesin family member 1B |
| chr14_+_50011161 | 0.33 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr18_-_5619309 | 0.32 |
ENSDART00000177705
|
CABZ01081638.1
|
ENSDARG00000107008 |
| chr5_+_63596281 | 0.30 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr18_-_321265 | 0.30 |
ENSDART00000136693
|
tm2d3
|
TM2 domain containing 3 |
| chr3_+_60035494 | 0.30 |
ENSDART00000166982
ENSDART00000166837 |
sec14l1
|
SEC14-like lipid binding 1 |
| chr5_-_1164454 | 0.30 |
ENSDART00000054070
|
surf2
|
surfeit 2 |
| chr13_-_12353560 | 0.30 |
ENSDART00000146195
|
lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr9_-_53274688 | 0.29 |
ENSDART00000140771
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr10_+_2848820 | 0.28 |
ENSDART00000141505
|
ccar2
|
cell cycle and apoptosis regulator 2 |
| chr20_+_12809388 | 0.27 |
ENSDART00000163499
|
zgc:153383
|
zgc:153383 |
| chr9_+_36023025 | 0.26 |
ENSDART00000122169
|
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr7_-_57680717 | 0.25 |
|
|
|
| chr9_+_14052160 | 0.25 |
ENSDART00000124267
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr25_+_31746760 | 0.24 |
ENSDART00000162636
|
tjp1b
|
tight junction protein 1b |
| chr2_+_42223071 | 0.23 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr4_+_11480865 | 0.22 |
ENSDART00000019458
|
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr7_-_6291359 | 0.22 |
ENSDART00000164372
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| KN150650v1_+_861 | 0.21 |
|
|
|
| chr3_-_10455344 | 0.18 |
ENSDART00000111833
|
pctp
|
phosphatidylcholine transfer protein |
| chr25_-_7801166 | 0.17 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr1_-_51086075 | 0.16 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr6_-_6097247 | 0.16 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr14_-_16118404 | 0.15 |
|
|
|
| chr11_-_17883113 | 0.15 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr21_+_2728070 | 0.15 |
|
|
|
| chr7_+_61450760 | 0.14 |
ENSDART00000165864
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr3_-_17722013 | 0.13 |
ENSDART00000074478
|
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr13_-_22568537 | 0.13 |
ENSDART00000016946
|
glud1a
|
glutamate dehydrogenase 1a |
| chr8_+_16722552 | 0.13 |
ENSDART00000142504
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr19_+_47707575 | 0.10 |
ENSDART00000163591
ENSDART00000159860 ENSDART00000138909 ENSDART00000158588 ENSDART00000170847 |
tpmt.2
|
thiopurine S-methyltransferase, tandem duplicate 2 |
| chr25_-_31952245 | 0.06 |
ENSDART00000153892
|
cep152
|
centrosomal protein 152 |
| chr25_-_7801200 | 0.06 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr18_+_38928197 | 0.05 |
|
|
|
| chr15_+_37688139 | 0.05 |
ENSDART00000076066
|
lin37
|
lin-37 DREAM MuvB core complex component |
| chr23_+_3570270 | 0.04 |
ENSDART00000092258
|
spata2
|
spermatogenesis associated 2 |
| chr1_+_37423518 | 0.01 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 0.9 | GO:1903798 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) small RNA loading onto RISC(GO:0070922) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.2 | 1.7 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.2 | 2.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 2.6 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.2 | 1.1 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 3.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 1.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 1.9 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 1.4 | GO:0031204 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 1.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.5 | GO:0070861 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.9 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 1.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 2.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.7 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.7 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 1.3 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 1.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.0 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 1.3 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 1.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.9 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.9 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.1 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.7 | GO:0006869 | lipid transport(GO:0006869) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 3.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.9 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.3 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 2.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 13.1 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.6 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 0.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.3 | 1.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 1.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.2 | 1.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0004353 | glutamate dehydrogenase (NAD+) activity(GO:0004352) glutamate dehydrogenase [NAD(P)+] activity(GO:0004353) oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.4 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |