Project
DANIO-CODE
Navigation
Downloads
Results for arid6+zgc:77151
Z-value: 0.95
Transcription factors associated with arid6+zgc:77151
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000054307 | 77151 |
|
arid6
|
ENSDARG00000069988 | AT-rich interaction domain 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arid6 | dr10_dc_chr21_+_11151424_11151440 | -0.36 | 1.7e-01 | Click! |
| zgc:77151 | dr10_dc_chr21_-_37341980_37342132 | -0.29 | 2.7e-01 | Click! |
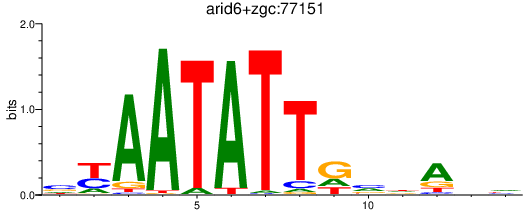
Activity profile of arid6+zgc:77151 motif
Sorted Z-values of arid6+zgc:77151 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arid6+zgc:77151
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_25734113 | 2.40 |
ENSDART00000136334
|
si:ch211-250e5.16
|
si:ch211-250e5.16 |
| chr15_-_17163526 | 1.79 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr12_+_13053552 | 1.68 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr21_-_7334721 | 1.39 |
ENSDART00000136671
|
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr23_+_370986 | 1.32 |
ENSDART00000055148
|
zgc:101663
|
zgc:101663 |
| chr13_-_18564182 | 1.32 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr20_-_14218080 | 1.30 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr12_+_26943407 | 1.18 |
ENSDART00000153054
|
fbrs
|
fibrosin |
| chr11_+_30400284 | 1.15 |
ENSDART00000169833
|
gb:eh507706
|
expressed sequence EH507706 |
| chr16_-_46426385 | 1.15 |
ENSDART00000141331
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr3_+_42383724 | 1.08 |
|
|
|
| chr20_+_14218237 | 1.00 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr10_+_33449922 | 0.97 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr7_+_13329491 | 0.96 |
ENSDART00000158797
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr24_+_40770039 | 0.94 |
|
|
|
| chr19_-_41820114 | 0.93 |
ENSDART00000038038
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr21_+_10485148 | 0.89 |
ENSDART00000165070
|
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr5_+_9579430 | 0.89 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr22_+_15317622 | 0.88 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr2_+_3115593 | 0.86 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr8_+_19945528 | 0.85 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr7_+_30355221 | 0.85 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr5_+_25408121 | 0.82 |
|
|
|
| chr16_+_32775164 | 0.82 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr8_-_1586037 | 0.82 |
ENSDART00000142703
|
CU694486.1
|
ENSDARG00000091948 |
| chr16_-_46558057 | 0.81 |
ENSDART00000169477
|
tmem176l.1
|
transmembrane protein 176l.1 |
| chr14_+_30058172 | 0.80 |
ENSDART00000053925
|
mtmr7a
|
myotubularin related protein 7a |
| chr19_+_24398149 | 0.78 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr17_+_17745119 | 0.77 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr20_+_48631807 | 0.76 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr10_+_33629943 | 0.73 |
ENSDART00000130093
|
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr16_+_28946580 | 0.73 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr15_+_24741931 | 0.73 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr17_-_31387062 | 0.73 |
|
|
|
| chr24_+_8702288 | 0.72 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr16_+_52961277 | 0.72 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr6_-_6951460 | 0.71 |
ENSDART00000148879
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr10_+_16071124 | 0.70 |
ENSDART00000065036
|
phax
|
phosphorylated adaptor for RNA export |
| chr5_-_69171508 | 0.69 |
ENSDART00000127782
|
ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr23_-_35691369 | 0.69 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr22_-_31089831 | 0.68 |
|
|
|
| chr6_+_12999546 | 0.68 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr2_-_48345919 | 0.68 |
|
|
|
| chr1_+_13719501 | 0.67 |
ENSDART00000030083
|
chic2
|
cysteine-rich hydrophobic domain 2 |
| chr20_+_14893184 | 0.66 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr16_+_25344257 | 0.66 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr17_+_21082384 | 0.66 |
ENSDART00000035432
|
entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr1_-_343261 | 0.65 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr21_+_40662814 | 0.65 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr10_+_24530670 | 0.65 |
|
|
|
| chr22_-_18466730 | 0.65 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr23_+_19728953 | 0.64 |
ENSDART00000104441
|
abhd6b
|
abhydrolase domain containing 6b |
| chr24_+_36429525 | 0.64 |
ENSDART00000062722
|
pus3
|
pseudouridylate synthase 3 |
| chr19_-_9020995 | 0.64 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr16_-_32773083 | 0.63 |
ENSDART00000137232
|
usp45
|
ubiquitin specific peptidase 45 |
| chr11_-_37613237 | 0.62 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr20_+_36725342 | 0.62 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr9_-_11884101 | 0.62 |
ENSDART00000146731
ENSDART00000134553 |
AL935144.1
|
ENSDARG00000093040 |
| chr5_+_58936399 | 0.61 |
|
|
|
| chr17_-_43542583 | 0.61 |
ENSDART00000125162
|
si:dkey-21a6.5
|
si:dkey-21a6.5 |
| chr8_-_4270732 | 0.61 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr19_+_24493786 | 0.60 |
ENSDART00000174798
|
BX548249.1
|
ENSDARG00000107081 |
| chr24_+_19270877 | 0.59 |
|
|
|
| chr13_+_24549364 | 0.59 |
ENSDART00000139854
|
zgc:66426
|
zgc:66426 |
| chr19_-_6274169 | 0.59 |
ENSDART00000140347
ENSDART00000092656 |
erf
|
Ets2 repressor factor |
| chr6_-_8009143 | 0.59 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr25_+_20596490 | 0.58 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr3_-_59690168 | 0.58 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr25_-_7490206 | 0.58 |
ENSDART00000163018
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr15_+_24741620 | 0.58 |
ENSDART00000078014
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr21_+_30684976 | 0.57 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr1_+_40428827 | 0.57 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr11_-_37816180 | 0.56 |
ENSDART00000086516
|
klhdc8a
|
kelch domain containing 8A |
| chr2_+_50680126 | 0.56 |
ENSDART00000122716
|
ENSDARG00000090398
|
ENSDARG00000090398 |
| chr14_+_35074248 | 0.56 |
ENSDART00000084914
|
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr2_-_47827141 | 0.56 |
ENSDART00000056882
|
cul3a
|
cullin 3a |
| chr17_+_32828254 | 0.56 |
|
|
|
| chr24_-_36383243 | 0.56 |
ENSDART00000155892
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr11_-_3516034 | 0.55 |
ENSDART00000009788
|
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr16_+_32775303 | 0.55 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr25_-_7546532 | 0.55 |
ENSDART00000131583
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr13_-_3805904 | 0.54 |
ENSDART00000017052
|
ncoa4
|
nuclear receptor coactivator 4 |
| chr16_+_52961219 | 0.54 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr16_+_29651573 | 0.54 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr21_+_13291305 | 0.53 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr9_+_2019992 | 0.53 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr15_+_20593440 | 0.53 |
ENSDART00000060935
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr20_-_14218236 | 0.53 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr21_-_39616469 | 0.53 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr12_+_20222000 | 0.52 |
ENSDART00000150020
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr20_-_21906340 | 0.52 |
ENSDART00000145807
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr17_-_23874858 | 0.52 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr13_-_37043725 | 0.52 |
|
|
|
| chr13_-_35766296 | 0.52 |
ENSDART00000162399
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_+_38959335 | 0.52 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr6_+_12999630 | 0.52 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr10_-_22967737 | 0.51 |
|
|
|
| chr10_+_26636293 | 0.51 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr15_+_36075070 | 0.51 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr6_-_3347207 | 0.50 |
ENSDART00000151416
|
si:ch1073-342h5.2
|
si:ch1073-342h5.2 |
| chr20_-_22631007 | 0.50 |
|
|
|
| chr5_-_47610384 | 0.50 |
|
|
|
| chr1_+_157840 | 0.50 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr16_-_28659489 | 0.49 |
ENSDART00000059053
|
rpp38
|
ribonuclease P/MRP 38 subunit |
| chr3_+_18657831 | 0.49 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr6_+_7165300 | 0.49 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr22_+_1153361 | 0.49 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr25_-_2263905 | 0.49 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr6_-_12434518 | 0.49 |
|
|
|
| chr15_+_36075206 | 0.48 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr16_+_25344184 | 0.48 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr2_-_32279019 | 0.48 |
ENSDART00000159843
ENSDART00000056621 ENSDART00000039717 ENSDART00000145704 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr5_+_25408083 | 0.48 |
|
|
|
| chr17_+_7377230 | 0.48 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr7_-_30654776 | 0.48 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr6_-_33893933 | 0.47 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr14_-_33517822 | 0.47 |
ENSDART00000112268
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr23_+_39454172 | 0.47 |
ENSDART00000149819
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr14_-_32525979 | 0.47 |
ENSDART00000166133
|
ube2a
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog) |
| chr18_+_21124516 | 0.47 |
ENSDART00000060191
|
suv420h1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr13_-_23888383 | 0.47 |
ENSDART00000026189
|
cog2
|
component of oligomeric golgi complex 2 |
| chr2_+_3115541 | 0.46 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr1_+_40428722 | 0.46 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr13_-_22776767 | 0.46 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| KN149945v1_-_7374 | 0.45 |
ENSDART00000160055
|
CABZ01048399.1
|
ENSDARG00000101896 |
| chr20_+_19093849 | 0.45 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr20_-_39886349 | 0.44 |
ENSDART00000098253
|
rnf217
|
ring finger protein 217 |
| chr13_+_46652067 | 0.44 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr24_-_25536050 | 0.44 |
ENSDART00000110241
|
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr21_-_27376521 | 0.44 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr13_-_23888408 | 0.43 |
ENSDART00000026189
|
cog2
|
component of oligomeric golgi complex 2 |
| chr21_+_20350218 | 0.43 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr17_+_21867968 | 0.43 |
ENSDART00000131929
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr12_+_47729417 | 0.43 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr7_+_20651600 | 0.43 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr5_-_65750231 | 0.43 |
ENSDART00000160189
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr13_-_28559160 | 0.43 |
ENSDART00000101582
|
pcgf6
|
polycomb group ring finger 6 |
| chr3_-_48458042 | 0.42 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr3_-_12199729 | 0.42 |
ENSDART00000165013
|
pam16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr19_-_10376361 | 0.42 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr25_-_34599241 | 0.42 |
ENSDART00000156727
|
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr14_-_15684734 | 0.42 |
ENSDART00000168075
|
trim105
|
tripartite motif containing 105 |
| KN150311v1_-_13973 | 0.42 |
ENSDART00000160072
|
snx1b
|
sorting nexin 1b |
| chr25_+_19772873 | 0.42 |
ENSDART00000170493
|
gramd4b
|
GRAM domain containing 4b |
| chr13_+_20742320 | 0.42 |
|
|
|
| chr17_-_29295450 | 0.42 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr15_+_15920149 | 0.41 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr14_+_20941526 | 0.41 |
ENSDART00000138551
|
smim19
|
small integral membrane protein 19 |
| chr15_-_17202268 | 0.41 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr9_-_41238226 | 0.40 |
ENSDART00000008275
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr25_-_19388381 | 0.40 |
ENSDART00000154986
|
zgc:193812
|
zgc:193812 |
| chr3_-_36277758 | 0.40 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr21_-_28304413 | 0.40 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr21_-_28304276 | 0.40 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr4_-_14981572 | 0.40 |
|
|
|
| chr25_+_29219116 | 0.40 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr5_-_45986774 | 0.40 |
|
|
|
| chr6_+_422911 | 0.40 |
|
|
|
| chr7_+_35163845 | 0.40 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr13_-_22776819 | 0.40 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr9_-_46614949 | 0.39 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr24_+_20782714 | 0.39 |
ENSDART00000144883
|
fam162a
|
family with sequence similarity 162, member A |
| chr21_+_40663230 | 0.39 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr3_-_31713730 | 0.39 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr14_+_42109615 | 0.39 |
|
|
|
| chr9_-_2522639 | 0.39 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr12_-_35481361 | 0.39 |
ENSDART00000158658
ENSDART00000168958 ENSDART00000162175 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr22_-_10026610 | 0.38 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr5_+_2440622 | 0.38 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr19_+_28707116 | 0.38 |
ENSDART00000103855
|
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr10_-_36747849 | 0.38 |
ENSDART00000122375
|
mrpl48
|
mitochondrial ribosomal protein L48 |
| chr16_-_41537827 | 0.38 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr20_+_30676086 | 0.38 |
ENSDART00000131422
ENSDART00000010494 |
fgfr1op
|
FGFR1 oncogene partner |
| chr22_+_22393190 | 0.38 |
ENSDART00000105600
|
ddx59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 |
| chr21_-_42872362 | 0.38 |
ENSDART00000171089
ENSDART00000160998 |
stk10
|
serine/threonine kinase 10 |
| chr2_+_38019379 | 0.37 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr3_-_31713951 | 0.37 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr2_-_37821146 | 0.37 |
ENSDART00000154124
|
nfatc4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr2_+_38019437 | 0.37 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr13_-_36486255 | 0.37 |
ENSDART00000160250
|
BX927407.1
|
ENSDARG00000103360 |
| chr12_+_46281592 | 0.37 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr5_-_34600655 | 0.37 |
ENSDART00000164396
|
fcho2
|
FCH domain only 2 |
| chr12_-_10438255 | 0.37 |
ENSDART00000106172
|
rac1a
|
ras-related C3 botulinum toxin substrate 1a (rho family, small GTP binding protein Rac1) |
| chr21_+_13291347 | 0.36 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr2_+_30439889 | 0.36 |
ENSDART00000139871
|
socs6b
|
suppressor of cytokine signaling 6b |
| chr22_+_699477 | 0.36 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr2_+_38019291 | 0.36 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr22_-_10026579 | 0.36 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr18_+_26061355 | 0.36 |
ENSDART00000015712
|
znf710a
|
zinc finger protein 710a |
| chr22_+_21593142 | 0.36 |
ENSDART00000133939
|
tle2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr13_-_28135403 | 0.35 |
ENSDART00000057565
|
sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr12_-_26943743 | 0.35 |
|
|
|
| chr18_+_44538877 | 0.35 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr9_+_34423285 | 0.35 |
ENSDART00000174944
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr20_+_48191608 | 0.35 |
ENSDART00000154114
|
BX005121.1
|
ENSDARG00000097548 |
| chr21_-_2597788 | 0.35 |
|
|
|
| chr14_+_51641126 | 0.35 |
ENSDART00000162321
|
cox18
|
COX18 cytochrome c oxidase assembly factor |
| chr16_-_25620064 | 0.35 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr18_+_31020260 | 0.34 |
ENSDART00000154993
|
cd151l
|
CD151 antigen, like |
| chr15_+_31920039 | 0.34 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 2.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 0.9 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.3 | 0.9 | GO:0042128 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.3 | 0.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 1.0 | GO:0010984 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.3 | 0.8 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.3 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.7 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.2 | 0.7 | GO:0098751 | bone cell development(GO:0098751) |
| 0.2 | 1.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.8 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 0.7 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 0.7 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.2 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.6 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of meiotic cell cycle(GO:0051447) |
| 0.1 | 0.8 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 1.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.3 | GO:0039528 | negative regulation of type I interferon production(GO:0032480) cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.1 | 0.4 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.3 | GO:0051701 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) interaction with host(GO:0051701) |
| 0.1 | 0.5 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.4 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.4 | GO:0050832 | neutrophil mediated immunity(GO:0002446) defense response to fungus(GO:0050832) |
| 0.1 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.3 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.3 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.3 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.5 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.3 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.9 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.2 | GO:0010481 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.2 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.4 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.0 | 0.8 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.7 | GO:0010324 | membrane invagination(GO:0010324) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.1 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 1.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 0.3 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.1 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.2 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0007052 | mitotic spindle organization(GO:0007052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 0.7 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 0.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 0.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.2 | GO:1990077 | primosome complex(GO:1990077) |
| 0.1 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 2.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.5 | 1.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.4 | 1.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 0.9 | GO:0008942 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.3 | 0.8 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.7 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.2 | 1.0 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 1.3 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 0.5 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.4 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.5 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 1.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.8 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 1.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.7 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |