Project
DANIO-CODE
Navigation
Downloads
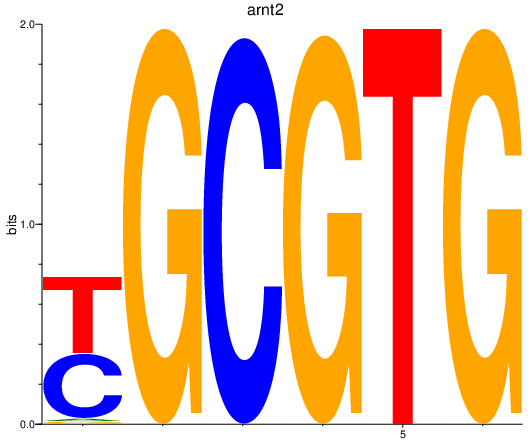
Results for arnt2
Z-value: 1.96
Transcription factors associated with arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arnt2
|
ENSDARG00000103697 | aryl-hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arnt2 | dr10_dc_chr7_+_10460031_10460135 | 0.76 | 6.6e-04 | Click! |
Activity profile of arnt2 motif
Sorted Z-values of arnt2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arnt2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_29277783 | 10.07 |
ENSDART00000149569
|
CR848047.1
|
ENSDARG00000095906 |
| chr21_-_20674965 | 4.91 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr7_+_49442972 | 4.83 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr14_+_35880163 | 4.47 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr16_+_24063849 | 4.13 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr9_+_25966225 | 4.05 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr14_+_47326080 | 3.98 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr21_+_6829161 | 3.96 |
ENSDART00000037265
|
olfm1b
|
olfactomedin 1b |
| chr5_-_47634182 | 3.91 |
ENSDART00000163250
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr8_-_11602501 | 3.89 |
ENSDART00000161743
|
fnbp1a
|
formin binding protein 1a |
| chr5_+_34397578 | 3.83 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr1_+_16681778 | 3.77 |
|
|
|
| chr21_+_17731439 | 3.74 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr16_+_49796978 | 3.69 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr15_+_38957922 | 3.67 |
ENSDART00000141086
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr8_+_29584750 | 3.51 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr7_-_14135850 | 3.31 |
ENSDART00000161442
|
CU467822.1
|
ENSDARG00000101584 |
| chr10_-_17527124 | 3.24 |
ENSDART00000137905
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr4_+_17428131 | 3.02 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr17_-_6580876 | 2.87 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr2_-_32370176 | 2.82 |
ENSDART00000077151
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr9_+_13149212 | 2.81 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr14_-_17257773 | 2.77 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr9_-_6683651 | 2.74 |
ENSDART00000061593
|
pou3f3a
|
POU class 3 homeobox 3a |
| chr5_+_49103778 | 2.70 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr23_+_20936419 | 2.70 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr5_+_48031920 | 2.66 |
ENSDART00000008043
ENSDART00000171438 |
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr9_-_44493074 | 2.55 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr23_+_14834670 | 2.51 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr20_+_35479511 | 2.50 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr9_+_33167554 | 2.50 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr4_-_12008472 | 2.49 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr5_-_11723101 | 2.48 |
ENSDART00000161706
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr23_+_20936374 | 2.45 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr6_+_42821679 | 2.44 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr6_-_22227089 | 2.43 |
ENSDART00000168554
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr20_-_8431338 | 2.42 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_+_6551983 | 2.41 |
ENSDART00000160382
ENSDART00000171495 ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr23_-_26150495 | 2.39 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr11_+_10925475 | 2.39 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr12_+_24221087 | 2.38 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr17_+_22936598 | 2.37 |
ENSDART00000155145
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr7_-_15582208 | 2.35 |
ENSDART00000102363
|
rcn1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr9_-_20562496 | 2.34 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr17_+_21944679 | 2.31 |
ENSDART00000063704
|
crip3
|
cysteine-rich protein 3 |
| chr23_-_6707246 | 2.27 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr17_+_17841882 | 2.27 |
ENSDART00000123311
|
ism2a
|
isthmin 2a |
| chr20_-_8431368 | 2.27 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr5_-_71539823 | 2.25 |
ENSDART00000167872
|
tbx3a
|
T-box 3a |
| chr9_+_25966366 | 2.20 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr23_-_31585883 | 2.19 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr7_+_55332040 | 2.19 |
ENSDART00000171736
|
trappc2l
|
trafficking protein particle complex 2-like |
| chr23_+_13911019 | 2.18 |
ENSDART00000168545
|
zbtb46
|
zinc finger and BTB domain containing 46 |
| chr18_-_44135616 | 2.15 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr4_+_8531280 | 2.14 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr8_-_47161379 | 2.10 |
ENSDART00000024320
|
ybx1
|
Y box binding protein 1 |
| chr21_+_23916512 | 2.07 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr8_+_19642272 | 2.05 |
ENSDART00000138176
|
foxd2
|
forkhead box D2 |
| chr2_+_51866323 | 2.05 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr9_-_9693163 | 2.01 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr11_-_24190842 | 2.00 |
ENSDART00000103752
|
dvl1b
|
dishevelled segment polarity protein 1b |
| chr5_-_34397435 | 2.00 |
ENSDART00000170684
|
btf3
|
basic transcription factor 3 |
| chr11_+_30569525 | 1.99 |
|
|
|
| chr24_-_21324086 | 1.96 |
|
|
|
| chr18_-_23889256 | 1.96 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_-_18868986 | 1.96 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr14_-_26406720 | 1.95 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr15_-_18638643 | 1.93 |
ENSDART00000142010
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr8_-_34077790 | 1.92 |
|
|
|
| chr9_-_20562293 | 1.91 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr13_+_17333810 | 1.91 |
ENSDART00000134181
|
CT025897.1
|
ENSDARG00000093577 |
| chr16_+_23172295 | 1.91 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr22_+_528076 | 1.88 |
|
|
|
| chr20_+_35479428 | 1.87 |
ENSDART00000159483
ENSDART00000031091 |
BX511024.1
vsnl1a
|
ENSDARG00000104812 visinin-like 1a |
| chr16_+_19926776 | 1.86 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr21_+_28921734 | 1.86 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_44942144 | 1.85 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr12_+_25684420 | 1.84 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr17_+_22359660 | 1.83 |
ENSDART00000162670
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr23_+_43284714 | 1.82 |
|
|
|
| chr4_+_18974767 | 1.82 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr17_+_38314814 | 1.81 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr17_+_34854673 | 1.80 |
|
|
|
| chr23_+_17294623 | 1.77 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr17_+_52736844 | 1.77 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr9_-_12840621 | 1.77 |
ENSDART00000088042
|
myo10l3
|
myosin X-like 3 |
| chr16_+_16941228 | 1.76 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr19_+_34145030 | 1.76 |
ENSDART00000151521
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_+_41523935 | 1.73 |
ENSDART00000110860
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr4_-_25080470 | 1.70 |
ENSDART00000179640
|
gata3
|
GATA binding protein 3 |
| chr16_+_50189177 | 1.70 |
ENSDART00000163565
|
plcl2
|
phospholipase C-like 2 |
| chr7_+_40604000 | 1.70 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr11_-_37322205 | 1.69 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr5_+_34397618 | 1.68 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr14_+_22150325 | 1.68 |
ENSDART00000108585
ENSDART00000161776 |
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr18_-_49121584 | 1.68 |
ENSDART00000174157
|
ENSDARG00000099137
|
ENSDARG00000099137 |
| chr20_+_34423970 | 1.67 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr2_+_29727235 | 1.66 |
ENSDART00000021987
|
ptf1a
|
pancreas specific transcription factor, 1a |
| chr12_+_35018663 | 1.66 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr11_-_28367271 | 1.65 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr7_+_32756620 | 1.64 |
|
|
|
| chr24_-_38068071 | 1.62 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr19_+_30800672 | 1.60 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr14_+_503643 | 1.60 |
|
|
|
| chr14_+_7626822 | 1.59 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr6_-_26569321 | 1.58 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr5_+_15319430 | 1.58 |
ENSDART00000162003
|
hspb8
|
heat shock protein b8 |
| chr23_+_31181140 | 1.58 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr1_-_22144014 | 1.56 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr17_+_52736535 | 1.54 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr6_+_33261144 | 1.54 |
ENSDART00000157428
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr12_-_26760324 | 1.53 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr13_-_20387917 | 1.53 |
ENSDART00000167916
ENSDART00000165310 ENSDART00000168955 |
gfra1a
|
gdnf family receptor alpha 1a |
| chr1_-_44942047 | 1.52 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr21_+_6858073 | 1.51 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr19_-_28546417 | 1.49 |
ENSDART00000130922
ENSDART00000079114 |
irx1b
|
iroquois homeobox 1b |
| chr17_-_2513630 | 1.49 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr6_-_39009073 | 1.49 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr20_-_20412830 | 1.48 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr11_+_41801746 | 1.47 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr19_+_17355544 | 1.46 |
ENSDART00000078951
|
MANEAL
|
mannosidase endo-alpha like |
| chr24_+_10276926 | 1.45 |
ENSDART00000145771
ENSDART00000157350 |
CT573344.1
|
ENSDARG00000093108 |
| chr2_-_43997672 | 1.45 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr25_-_7635824 | 1.45 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr5_+_35815450 | 1.44 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr11_-_27455242 | 1.44 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr16_-_23882488 | 1.44 |
ENSDART00000077834
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr10_+_5693933 | 1.43 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr6_+_11594333 | 1.43 |
ENSDART00000109552
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr13_-_37527812 | 1.43 |
ENSDART00000143806
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr23_+_17853977 | 1.42 |
|
|
|
| chr25_-_35539585 | 1.41 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr9_-_23406315 | 1.41 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr21_+_17073163 | 1.39 |
|
|
|
| chr19_-_10324632 | 1.39 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr12_-_1544844 | 1.38 |
|
|
|
| chr3_-_33773025 | 1.38 |
|
|
|
| chr2_+_51087570 | 1.37 |
|
|
|
| chr23_-_38228305 | 1.37 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr8_-_19872014 | 1.37 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr10_-_35293024 | 1.37 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr18_-_23889025 | 1.36 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_39491659 | 1.36 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr23_-_31629162 | 1.36 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr9_-_24602136 | 1.35 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr23_+_13367978 | 1.34 |
ENSDART00000139475
|
samd10b
|
sterile alpha motif domain containing 10b |
| chr7_+_19765237 | 1.32 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr21_-_485916 | 1.32 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr13_+_26943081 | 1.31 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr11_-_27455348 | 1.30 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr19_-_43081969 | 1.30 |
ENSDART00000123360
|
susd5
|
sushi domain containing 5 |
| chr12_-_37921731 | 1.30 |
|
|
|
| chr6_-_39008833 | 1.30 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr19_+_19839981 | 1.29 |
ENSDART00000113580
|
ENSDARG00000074216
|
ENSDARG00000074216 |
| chr15_-_45446153 | 1.29 |
ENSDART00000100332
|
fgf12b
|
fibroblast growth factor 12b |
| chr25_+_24193604 | 1.29 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr1_-_50066633 | 1.29 |
|
|
|
| chr9_+_36316158 | 1.28 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr7_-_42972804 | 1.28 |
ENSDART00000054552
|
cdh8
|
cadherin 8 |
| chr4_+_7879334 | 1.28 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr2_-_24947660 | 1.27 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr24_+_23646663 | 1.26 |
ENSDART00000146580
|
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr18_-_39797167 | 1.25 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr11_+_34562255 | 1.25 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
| chr24_+_40292617 | 1.25 |
|
|
|
| chr18_-_23888988 | 1.25 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr11_+_7314878 | 1.24 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr11_-_12067922 | 1.24 |
ENSDART00000110117
|
socs7
|
suppressor of cytokine signaling 7 |
| chr12_-_4597181 | 1.23 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr19_+_21783111 | 1.23 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr10_-_26782374 | 1.23 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr10_-_2655144 | 1.22 |
|
|
|
| chr15_-_31635737 | 1.22 |
ENSDART00000156047
|
hmgb1b
|
high mobility group box 1b |
| chr7_-_33080261 | 1.22 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr20_+_32646617 | 1.22 |
|
|
|
| chr9_-_44487895 | 1.21 |
ENSDART00000110411
|
cerkl
|
ceramide kinase-like |
| chr21_+_6829352 | 1.21 |
ENSDART00000146371
|
olfm1b
|
olfactomedin 1b |
| chr12_+_25684362 | 1.20 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr24_+_37518715 | 1.19 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr2_-_42643045 | 1.19 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr8_+_20592039 | 1.19 |
ENSDART00000160421
|
nfic
|
nuclear factor I/C |
| chr11_+_3940085 | 1.18 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr1_-_22066408 | 1.18 |
ENSDART00000134719
ENSDART00000166891 |
prom1b
|
prominin 1 b |
| chr23_-_45356039 | 1.17 |
ENSDART00000067630
|
ENSDARG00000039901
|
ENSDARG00000039901 |
| chr8_-_19872129 | 1.17 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr7_-_33678058 | 1.16 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr14_+_16508093 | 1.16 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr5_+_31562242 | 1.15 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr17_-_23947646 | 1.15 |
|
|
|
| chr12_+_35018567 | 1.15 |
ENSDART00000164597
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr23_-_1124499 | 1.15 |
|
|
|
| chr18_-_23889169 | 1.14 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_22047445 | 1.14 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr5_-_40961381 | 1.14 |
|
|
|
| chr22_-_26971466 | 1.13 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr3_+_17947837 | 1.13 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr2_+_31974269 | 1.13 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr5_-_34393017 | 1.12 |
ENSDART00000134516
|
btf3
|
basic transcription factor 3 |
| chr6_-_14162846 | 1.12 |
ENSDART00000100762
|
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr15_+_28550096 | 1.12 |
ENSDART00000154320
|
ankrd13b
|
ankyrin repeat domain 13B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 1.2 | 4.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 1.1 | 5.7 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.1 | 4.5 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 1.0 | 6.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 1.0 | 3.0 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 1.0 | 3.0 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 1.0 | 3.0 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.9 | 3.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.9 | 2.7 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.9 | 2.7 | GO:0030815 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.9 | 3.5 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.8 | 2.5 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.7 | 2.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.6 | 2.4 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) negative regulation of cell aging(GO:0090344) regulation of cellular senescence(GO:2000772) |
| 0.6 | 2.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.6 | 1.7 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.5 | 3.1 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.5 | 2.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 1.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.5 | 1.4 | GO:0001823 | ureteric bud development(GO:0001657) branching involved in ureteric bud morphogenesis(GO:0001658) mesonephros development(GO:0001823) negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) ureteric bud morphogenesis(GO:0060675) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) mesonephric tubule morphogenesis(GO:0072171) |
| 0.4 | 1.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 1.2 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.4 | 1.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.4 | 1.4 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.3 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 1.7 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.3 | 3.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 2.3 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 1.0 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.3 | 1.3 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.3 | 0.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.3 | 2.0 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.3 | 1.7 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.3 | 2.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.3 | 1.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 | 0.8 | GO:0030431 | sleep(GO:0030431) |
| 0.3 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.3 | 0.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.3 | 1.3 | GO:0015817 | histidine transport(GO:0015817) |
| 0.2 | 1.9 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.2 | 0.7 | GO:0051701 | interaction with host(GO:0051701) |
| 0.2 | 0.7 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.2 | 1.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 0.9 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.2 | 1.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 0.9 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 3.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 0.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 1.0 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 0.8 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) |
| 0.2 | 1.0 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 1.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.7 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.2 | 1.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.2 | 1.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 4.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 0.4 | GO:2000136 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.1 | 1.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 8.6 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 3.0 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.8 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.3 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.4 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.1 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 1.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 1.8 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 4.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 3.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.3 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.1 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.6 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 3.2 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 0.8 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 4.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.1 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 0.4 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 1.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.0 | GO:0003208 | cardiac ventricle morphogenesis(GO:0003208) |
| 0.1 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.4 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 3.0 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 3.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 1.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 1.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.0 | 1.6 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.2 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 2.6 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 2.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.3 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 1.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.8 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.2 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 1.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.7 | 5.1 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.6 | 2.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.5 | 2.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.5 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 1.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 1.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 1.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 1.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 2.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 3.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 10.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 19.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.9 | GO:0030133 | transport vesicle(GO:0030133) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.8 | 2.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.7 | 3.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 5.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 1.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.4 | 1.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.4 | 3.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 1.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 2.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 1.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.4 | 1.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 1.4 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.4 | 1.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.4 | 1.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 1.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 1.2 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.3 | 1.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 1.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 1.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 2.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 2.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 2.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 2.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 3.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 2.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 2.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 2.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 2.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 1.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 0.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 2.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 1.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 1.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 2.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 5.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 4.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.7 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 2.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 3.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 2.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 3.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 26.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 39.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 4.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 3.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0019869 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) neuropeptide binding(GO:0042923) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 11.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 2.7 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 8.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 1.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 3.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 3.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 2.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 2.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 3.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 0.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 3.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 6.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.4 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |