Project
DANIO-CODE
Navigation
Downloads
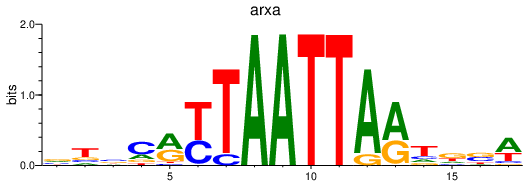
Results for arxa
Z-value: 1.08
Transcription factors associated with arxa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
arxa
|
ENSDARG00000058011 | aristaless related homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| arxa | dr10_dc_chr24_-_23797468_23797551 | -0.66 | 5.6e-03 | Click! |
Activity profile of arxa motif
Sorted Z-values of arxa motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arxa
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37303599 | 4.15 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr17_-_45021393 | 3.94 |
|
|
|
| chr7_-_24249672 | 2.75 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr18_-_40718244 | 2.70 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr6_+_36862078 | 2.69 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr21_+_6289363 | 2.39 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr2_-_26941084 | 2.17 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr14_-_6738260 | 2.05 |
ENSDART00000171792
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr24_+_19270877 | 1.98 |
|
|
|
| chr8_-_53165501 | 1.97 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr18_-_43890836 | 1.88 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr20_-_29961498 | 1.79 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr20_-_29961589 | 1.77 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr2_+_56534374 | 1.73 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr11_-_38856849 | 1.69 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr4_-_3340315 | 1.69 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr14_+_34218144 | 1.68 |
ENSDART00000075170
|
pttg1
|
pituitary tumor-transforming 1 |
| chr7_-_24249630 | 1.59 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr1_+_35253862 | 1.58 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr2_-_17721810 | 1.53 |
ENSDART00000100201
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr11_-_2437396 | 1.51 |
|
|
|
| chr2_-_26941232 | 1.46 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr24_-_14446593 | 1.46 |
|
|
|
| chr23_-_19299279 | 1.36 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr18_-_14306241 | 1.35 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr21_+_13291305 | 1.34 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr1_+_46474561 | 1.33 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr3_+_17783319 | 1.32 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr10_-_17214368 | 1.29 |
|
|
|
| chr14_-_14353487 | 1.28 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr15_+_35089305 | 1.26 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr1_+_46808632 | 1.25 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr2_-_26940965 | 1.20 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr25_-_1243081 | 1.19 |
ENSDART00000156062
|
calml4b
|
calmodulin-like 4b |
| chr6_+_36862148 | 1.17 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr12_-_33256671 | 1.17 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr13_+_12629211 | 1.16 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr14_-_49777408 | 1.14 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr19_+_1743359 | 1.11 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr20_+_29306677 | 1.11 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr7_-_26226924 | 1.09 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr15_+_34734212 | 1.08 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr21_+_13291242 | 1.08 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr6_+_36861678 | 1.07 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr18_+_39125694 | 1.07 |
ENSDART00000122377
|
bcl2l10
|
BCL2-like 10 (apoptosis facilitator) |
| chr19_-_1571878 | 1.06 |
|
|
|
| chr5_+_37303707 | 1.06 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr13_-_36672424 | 1.06 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr16_-_28723759 | 1.06 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr21_-_15019953 | 1.02 |
|
|
|
| chr16_+_23884575 | 1.01 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr12_-_33256754 | 1.01 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_+_35153749 | 1.00 |
ENSDART00000151073
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr22_-_16416848 | 0.98 |
ENSDART00000006290
|
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr3_+_40022244 | 0.97 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr16_-_55263338 | 0.97 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr15_+_31709362 | 0.97 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr25_+_7067115 | 0.97 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr20_+_27341244 | 0.97 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr6_+_40924559 | 0.94 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr23_+_28464194 | 0.94 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr25_+_7067043 | 0.93 |
ENSDART00000179500
|
BX005076.1
|
ENSDARG00000107609 |
| chr14_-_33141111 | 0.92 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr25_-_20933295 | 0.92 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr12_+_33257120 | 0.91 |
|
|
|
| chr11_+_40847576 | 0.91 |
ENSDART00000041531
|
park7
|
parkinson protein 7 |
| chr1_+_8468971 | 0.90 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr7_-_26226628 | 0.90 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr20_+_29306863 | 0.89 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_-_3673698 | 0.89 |
|
|
|
| chr21_-_44086464 | 0.88 |
ENSDART00000130833
|
FO704810.1
|
ENSDARG00000087492 |
| chr20_-_37910887 | 0.88 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr9_-_11884101 | 0.88 |
ENSDART00000146731
ENSDART00000134553 |
AL935144.1
|
ENSDARG00000093040 |
| chr21_+_13291347 | 0.88 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr2_+_1645259 | 0.87 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr21_-_45323151 | 0.86 |
ENSDART00000151106
|
ENSDARG00000053404
|
ENSDARG00000053404 |
| chr5_+_31475897 | 0.85 |
ENSDART00000144510
|
zmat5
|
zinc finger, matrin-type 5 |
| chr3_+_19490148 | 0.85 |
|
|
|
| chr23_-_19299398 | 0.84 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr1_+_18740188 | 0.81 |
ENSDART00000144403
|
clockb
|
clock circadian regulator b |
| chr19_-_2084484 | 0.78 |
|
|
|
| chr12_-_33257026 | 0.77 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_+_26040518 | 0.76 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr6_+_40925259 | 0.76 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr19_+_5562075 | 0.76 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr22_-_7887341 | 0.75 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr2_+_11094785 | 0.75 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr12_-_33256934 | 0.74 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr25_-_13394261 | 0.73 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr16_-_30476223 | 0.73 |
ENSDART00000018504
|
zgc:77086
|
zgc:77086 |
| chr12_-_33256599 | 0.72 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr15_+_31709330 | 0.72 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr7_+_23752492 | 0.71 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr13_+_12629243 | 0.67 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr21_-_30045669 | 0.66 |
ENSDART00000155188
|
ccnjl
|
cyclin J-like |
| chr23_+_28464642 | 0.65 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr9_-_35127938 | 0.65 |
ENSDART00000077814
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr9_+_24255311 | 0.65 |
ENSDART00000166303
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr18_-_20605032 | 0.63 |
ENSDART00000134722
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr4_-_1952230 | 0.63 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr23_+_28464298 | 0.63 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr20_+_20831223 | 0.63 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr20_+_25687135 | 0.62 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr6_-_40924459 | 0.62 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr17_+_5454020 | 0.61 |
ENSDART00000097455
|
fam167aa
|
family with sequence similarity 167, member Aa |
| chr22_-_7887401 | 0.61 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr16_+_19223683 | 0.60 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr2_+_10209233 | 0.60 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_-_20933145 | 0.60 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr11_-_2437361 | 0.60 |
|
|
|
| chr20_-_22532817 | 0.57 |
ENSDART00000177772
|
BX276097.1
|
ENSDARG00000108228 |
| chr8_-_17132057 | 0.57 |
ENSDART00000143920
|
mrps36
|
mitochondrial ribosomal protein S36 |
| chr4_-_5100307 | 0.57 |
ENSDART00000138817
|
tmem209
|
transmembrane protein 209 |
| chr11_+_16017857 | 0.56 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_-_73204361 | 0.56 |
ENSDART00000157500
|
AL929105.1
|
ENSDARG00000105194 |
| chr12_+_3632548 | 0.55 |
|
|
|
| chr7_-_5253629 | 0.54 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr9_+_2523927 | 0.54 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr5_-_11309164 | 0.53 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr20_+_27341324 | 0.53 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr20_+_35176069 | 0.52 |
|
|
|
| chr21_+_43333462 | 0.52 |
ENSDART00000109620
|
sept8a
|
septin 8a |
| chr2_-_47827343 | 0.51 |
ENSDART00000056882
|
cul3a
|
cullin 3a |
| chr1_+_51087450 | 0.50 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr13_-_36672285 | 0.50 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr6_+_49489473 | 0.49 |
ENSDART00000177749
|
FO704848.1
|
ENSDARG00000107438 |
| chr20_-_29961621 | 0.49 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr20_+_38555185 | 0.49 |
ENSDART00000020153
|
adck3
|
aarF domain containing kinase 3 |
| chr24_+_16402587 | 0.49 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr20_+_29306945 | 0.48 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr16_-_31386496 | 0.47 |
|
|
|
| chr20_+_40560443 | 0.45 |
ENSDART00000153119
|
serinc1
|
serine incorporator 1 |
| chr2_+_10192670 | 0.45 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_-_10694465 | 0.44 |
ENSDART00000127054
|
BX572619.1
|
ENSDARG00000089303 |
| chr1_+_46474845 | 0.43 |
ENSDART00000053152
|
cbr1
|
carbonyl reductase 1 |
| chr7_-_29906449 | 0.43 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr6_+_49552893 | 0.42 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr5_-_51363461 | 0.42 |
ENSDART00000051003
|
cdk7
|
cyclin-dependent kinase 7 |
| chr16_-_28723722 | 0.40 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr7_+_21006509 | 0.39 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr2_+_47827350 | 0.38 |
|
|
|
| chr2_-_11094083 | 0.37 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr10_-_2761480 | 0.36 |
ENSDART00000131749
|
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr23_+_34120812 | 0.33 |
ENSDART00000143933
|
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr12_-_308764 | 0.33 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr3_+_32234155 | 0.33 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr24_+_37752868 | 0.32 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr20_+_22900165 | 0.31 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr7_+_23752651 | 0.31 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr21_+_10757086 | 0.30 |
|
|
|
| chr4_-_3340275 | 0.30 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr16_-_31393328 | 0.29 |
ENSDART00000178298
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr14_+_37317006 | 0.29 |
|
|
|
| chr22_-_36561217 | 0.29 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr24_-_34794856 | 0.29 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr11_+_35790782 | 0.29 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
| chr25_+_29687993 | 0.28 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr17_-_41000670 | 0.28 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr14_-_470112 | 0.28 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr2_+_35824136 | 0.27 |
ENSDART00000137384
|
rasal2
|
RAS protein activator like 2 |
| chr6_+_3313515 | 0.27 |
ENSDART00000011785
|
rad54l
|
RAD54-like (S. cerevisiae) |
| chr16_-_13927947 | 0.26 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr20_+_22900302 | 0.25 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr16_+_19223553 | 0.24 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr4_-_1952201 | 0.23 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr3_-_23442774 | 0.23 |
|
|
|
| chr16_+_51597409 | 0.22 |
|
|
|
| chr3_-_32686757 | 0.22 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr21_+_43177310 | 0.21 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr14_+_10648299 | 0.21 |
ENSDART00000145776
|
zdhhc15b
|
zinc finger, DHHC-type containing 15b |
| chr17_-_36904594 | 0.21 |
|
|
|
| chr15_-_36490729 | 0.19 |
|
|
|
| chr7_+_22272746 | 0.19 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr19_-_2084281 | 0.19 |
|
|
|
| chr14_-_6831278 | 0.18 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr17_-_16414629 | 0.17 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr8_+_7740024 | 0.16 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr12_+_35849423 | 0.16 |
|
|
|
| chr17_+_8642686 | 0.15 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr16_-_31664756 | 0.14 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr5_-_67395562 | 0.13 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr2_+_50873843 | 0.13 |
|
|
|
| chr9_+_29832294 | 0.13 |
ENSDART00000023210
|
trim13
|
tripartite motif containing 13 |
| chr24_-_14446522 | 0.12 |
|
|
|
| chr22_-_4867538 | 0.11 |
ENSDART00000081801
|
ncl1
|
nicalin |
| chr17_+_43041951 | 0.11 |
ENSDART00000154863
|
isca2
|
iron-sulfur cluster assembly 2 |
| chr2_-_10192027 | 0.11 |
ENSDART00000100709
|
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr12_+_22459371 | 0.10 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| KN149711v1_-_7827 | 0.10 |
ENSDART00000171410
|
ENSDARG00000104905
|
ENSDARG00000104905 |
| chr1_+_35253821 | 0.09 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr6_+_25161456 | 0.09 |
ENSDART00000157790
|
ccbl2
|
cysteine conjugate-beta lyase 2 |
| chr16_+_54350976 | 0.09 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr24_-_37752716 | 0.08 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr1_+_18739982 | 0.06 |
ENSDART00000144403
|
clockb
|
clock circadian regulator b |
| chr21_-_45322936 | 0.06 |
ENSDART00000151106
|
ENSDARG00000053404
|
ENSDARG00000053404 |
| chr9_+_24255064 | 0.06 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr12_-_309047 | 0.06 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr20_+_29307283 | 0.05 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr10_+_40293158 | 0.05 |
|
|
|
| chr10_+_40293111 | 0.05 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 1.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.3 | 1.0 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.3 | 1.8 | GO:0036268 | swimming(GO:0036268) |
| 0.3 | 0.9 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.3 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.3 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.9 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.7 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.1 | 1.7 | GO:2000816 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 1.3 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 4.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 2.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.1 | GO:0051279 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 4.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 1.0 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.5 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.8 | GO:0006183 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.1 | 1.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.0 | 0.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.8 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 2.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 2.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.3 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 4.3 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 2.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 0.9 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 0.9 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.3 | 1.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.0 | GO:0032593 | lateral plasma membrane(GO:0016328) insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 4.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 1.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.2 | 1.5 | GO:0015562 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 2.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 5.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.2 | 1.0 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 1.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 4.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.3 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 4.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 4.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.8 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 2.3 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |