Project
DANIO-CODE
Navigation
Downloads
Results for arxb_drgx
Z-value: 1.26
Transcription factors associated with arxb_drgx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
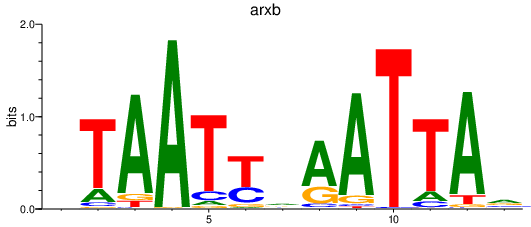
arxb
|
ENSDARG00000101523 | aristaless related homeobox b |
|
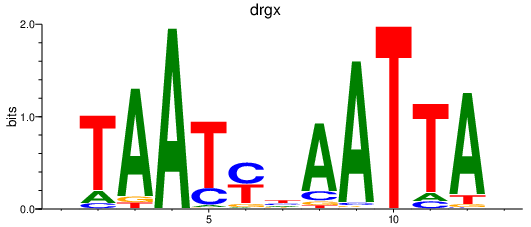
drgx
|
ENSDARG00000069329 | dorsal root ganglia homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| drgx | dr10_dc_chr13_+_30781652_30781657 | 0.68 | 3.6e-03 | Click! |
| arxb | dr10_dc_chr21_-_40653250_40653254 | 0.59 | 1.6e-02 | Click! |
Activity profile of arxb_drgx motif
Sorted Z-values of arxb_drgx motif
Network of associatons between targets according to the STRING database.
First level regulatory network of arxb_drgx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_18866796 | 5.83 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr7_+_21593005 | 4.43 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr1_+_44205158 | 4.04 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr6_+_16341787 | 3.80 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr22_-_32553488 | 3.08 |
ENSDART00000104693
|
pcbp4
|
poly(rC) binding protein 4 |
| chr16_-_31763183 | 2.75 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr7_+_26357875 | 2.72 |
ENSDART00000101044
|
hsbp1a
|
heat shock factor binding protein 1a |
| chr4_+_9668755 | 2.64 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr14_-_20760787 | 2.51 |
|
|
|
| chr6_+_23787804 | 2.43 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr6_-_15444945 | 2.31 |
|
|
|
| chr20_-_21031504 | 2.27 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr16_+_24069711 | 2.11 |
ENSDART00000142869
|
apoc2
|
apolipoprotein C-II |
| chr21_+_7844259 | 1.98 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr2_+_30932612 | 1.90 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr4_+_5826813 | 1.72 |
ENSDART00000055414
|
pex26
|
peroxisomal biogenesis factor 26 |
| chr6_+_16342020 | 1.72 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr7_-_30096283 | 1.63 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr1_-_40208544 | 1.62 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr7_-_27877036 | 1.60 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr17_-_19325678 | 1.59 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr5_-_30324182 | 1.56 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr12_+_14702847 | 1.54 |
|
|
|
| chr16_-_21334269 | 1.51 |
ENSDART00000145837
|
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr1_+_13779755 | 1.48 |
|
|
|
| chr21_-_27845076 | 1.43 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr10_-_26782374 | 1.39 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr2_-_48103519 | 1.39 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr17_-_8111181 | 1.36 |
ENSDART00000149873
ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr22_-_15567180 | 1.34 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr23_-_18203680 | 1.34 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr13_-_31491759 | 1.33 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr1_-_40208469 | 1.29 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr18_-_13153360 | 1.27 |
ENSDART00000092648
|
tmem5
|
transmembrane protein 5 |
| chr12_+_42281025 | 1.27 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr7_+_19895162 | 1.26 |
ENSDART00000011398
|
si:ch73-335l21.1
|
si:ch73-335l21.1 |
| chr2_+_27344633 | 1.26 |
ENSDART00000178275
|
cdh7
|
cadherin 7, type 2 |
| chr11_-_44724371 | 1.25 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr6_+_43236843 | 1.21 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr5_+_48031920 | 1.19 |
ENSDART00000008043
ENSDART00000171438 |
adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr14_-_47083485 | 1.16 |
ENSDART00000172512
|
elf2a
|
E74-like factor 2a (ets domain transcription factor) |
| chr24_+_19374200 | 1.14 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr9_+_34832049 | 1.10 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr24_-_38496534 | 1.03 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr18_+_23891068 | 1.00 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr21_-_44109455 | 0.99 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr13_-_29290894 | 0.97 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr5_+_50432868 | 0.95 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr9_-_51975919 | 0.93 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr23_-_24755654 | 0.90 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr2_-_49114158 | 0.90 |
|
|
|
| chr24_-_23797468 | 0.86 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr2_-_42222069 | 0.86 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr24_+_19374099 | 0.82 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr14_-_16791447 | 0.81 |
ENSDART00000158002
|
CR812832.1
|
ENSDARG00000103278 |
| chr25_+_4918339 | 0.80 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr6_+_6767424 | 0.77 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr21_-_28883441 | 0.75 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr14_-_24304373 | 0.75 |
|
|
|
| chr18_+_23891152 | 0.75 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr19_-_35739239 | 0.71 |
|
|
|
| chr3_-_26657213 | 0.69 |
|
|
|
| chr7_+_13753344 | 0.68 |
|
|
|
| chr6_+_8895437 | 0.66 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr23_-_12410797 | 0.65 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr12_+_17382681 | 0.65 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr14_-_16762869 | 0.64 |
ENSDART00000163766
|
phox2bb
|
paired-like homeobox 2bb |
| chr2_-_20490037 | 0.64 |
ENSDART00000160388
|
FQ377605.1
|
ENSDARG00000101927 |
| chr13_+_33378267 | 0.61 |
ENSDART00000025007
|
jag2a
|
jagged 2a |
| chr13_+_14845093 | 0.61 |
ENSDART00000011520
|
noto
|
notochord homeobox |
| chr23_-_10906094 | 0.60 |
|
|
|
| chr18_+_23890892 | 0.58 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr15_+_22499643 | 0.58 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr6_+_40664212 | 0.57 |
ENSDART00000103842
|
eno1b
|
enolase 1b, (alpha) |
| chr14_-_16762762 | 0.54 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr14_-_21854679 | 0.54 |
ENSDART00000115422
|
nudcd2
|
NudC domain containing 2 |
| chr24_+_30343717 | 0.53 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr16_-_31763281 | 0.53 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr3_+_3923243 | 0.52 |
ENSDART00000104977
|
pick1
|
protein interacting with prkca 1 |
| chr11_-_37158131 | 0.50 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr13_+_18376642 | 0.49 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr19_+_16160028 | 0.49 |
ENSDART00000144376
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr23_+_28564630 | 0.47 |
|
|
|
| chr23_+_5899604 | 0.42 |
|
|
|
| chr5_-_23709401 | 0.40 |
|
|
|
| chr12_-_34612821 | 0.39 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr13_-_49707003 | 0.39 |
|
|
|
| chr22_+_37688585 | 0.39 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr10_+_9259213 | 0.38 |
ENSDART00000064966
|
snx18b
|
sorting nexin 18b |
| chr20_-_29028920 | 0.38 |
ENSDART00000153082
|
susd6
|
sushi domain containing 6 |
| chr24_-_21750082 | 0.38 |
|
|
|
| chr7_+_71757218 | 0.38 |
ENSDART00000012918
|
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr14_-_20826302 | 0.37 |
ENSDART00000171679
|
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr13_+_49154065 | 0.37 |
|
|
|
| chr23_-_36342779 | 0.33 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr7_+_28896401 | 0.32 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr3_+_29314649 | 0.30 |
ENSDART00000020381
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr8_-_17130805 | 0.30 |
|
|
|
| chr24_+_30343687 | 0.30 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr24_+_24924379 | 0.29 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr1_+_45843326 | 0.28 |
|
|
|
| KN149962v1_-_109672 | 0.28 |
|
|
|
| chr17_-_12812747 | 0.27 |
ENSDART00000022874
ENSDART00000046025 |
psma6a
|
proteasome subunit alpha 6a |
| chr14_-_34265449 | 0.27 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr7_+_21592861 | 0.26 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr1_+_18118995 | 0.26 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr14_-_24113237 | 0.25 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr20_+_10179249 | 0.24 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr21_-_10354049 | 0.23 |
ENSDART00000159958
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr20_-_43878454 | 0.22 |
ENSDART00000100610
ENSDART00000100608 ENSDART00000148809 |
matn3a
|
matrilin 3a |
| chr20_+_31384580 | 0.22 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr3_-_30948297 | 0.22 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr7_+_38540519 | 0.22 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr3_-_13771322 | 0.21 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr24_-_17089437 | 0.20 |
|
|
|
| chr12_+_42281154 | 0.19 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr9_-_27838603 | 0.19 |
|
|
|
| chr1_+_37423689 | 0.18 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr13_+_46487890 | 0.18 |
|
|
|
| chr10_+_22538287 | 0.18 |
ENSDART00000109070
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr22_+_17409030 | 0.18 |
ENSDART00000139523
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_+_38540586 | 0.17 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr3_+_31047152 | 0.16 |
|
|
|
| chr21_-_28883243 | 0.15 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr19_-_35739013 | 0.12 |
|
|
|
| chr20_-_21031592 | 0.11 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr2_+_20946939 | 0.10 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr1_-_55195428 | 0.10 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr9_-_48709908 | 0.09 |
ENSDART00000058248
|
ENSDARG00000039824
|
ENSDARG00000039824 |
| chr14_+_6308806 | 0.09 |
ENSDART00000139292
|
si:dkeyp-44a8.2
|
si:dkeyp-44a8.2 |
| chr8_-_18167468 | 0.08 |
ENSDART00000137005
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr1_+_25110899 | 0.08 |
ENSDART00000129471
|
gucy1b3
|
guanylate cyclase 1, soluble, beta 3 |
| chr14_-_29518685 | 0.08 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr8_-_18167570 | 0.08 |
ENSDART00000142114
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr9_-_9727433 | 0.07 |
ENSDART00000149021
|
gpr156
|
G protein-coupled receptor 156 |
| chr9_-_51976213 | 0.07 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr15_+_8791540 | 0.06 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr1_+_37423518 | 0.06 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_+_52225276 | 0.05 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr2_+_29729161 | 0.05 |
ENSDART00000141666
|
AL845362.1
|
ENSDARG00000093205 |
| chr19_+_42658273 | 0.05 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr15_-_5732737 | 0.04 |
ENSDART00000109599
|
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr2_-_14938196 | 0.04 |
ENSDART00000170687
|
pde4bb
|
phosphodiesterase 4B, cAMP-specific b |
| chr14_-_21717949 | 0.04 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr5_+_31889674 | 0.03 |
ENSDART00000165417
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr22_-_15666856 | 0.03 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr5_-_68408171 | 0.03 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr10_-_33677866 | 0.02 |
ENSDART00000142655
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr20_-_43281081 | 0.01 |
|
|
|
| chr8_+_35483042 | 0.00 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr17_-_45140696 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.8 | 3.1 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.6 | 5.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.6 | 1.7 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.5 | 1.6 | GO:0061130 | branching involved in pancreas morphogenesis(GO:0061114) pancreatic bud formation(GO:0061130) pancreas field specification(GO:0061131) |
| 0.5 | 2.0 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.4 | 1.2 | GO:0030818 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.4 | 1.2 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 1.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 1.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 2.9 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 1.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 1.3 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 1.6 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.2 | 2.0 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 0.7 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) regulation of calcium-mediated signaling(GO:0050848) |
| 0.2 | 2.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.6 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 0.5 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.1 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 1.1 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.7 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) stereocilium(GO:0032420) |
| 0.2 | 1.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.5 | 3.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 1.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.3 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.2 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 2.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.9 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 0.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 16.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |