Project
DANIO-CODE
Navigation
Downloads
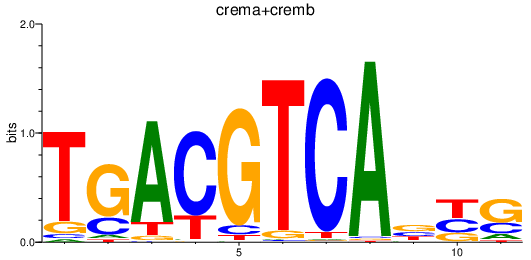
Results for atf2_atf1_atf3_crema+cremb
Z-value: 3.06
Transcription factors associated with atf2_atf1_atf3_crema+cremb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
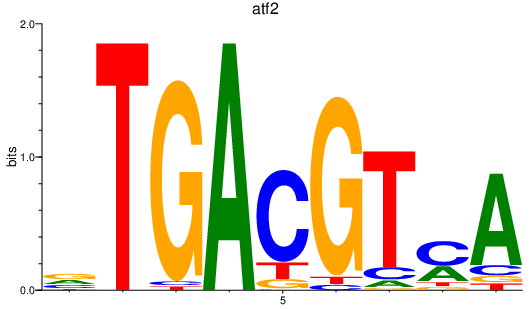
atf2
|
ENSDARG00000023903 | activating transcription factor 2 |
|
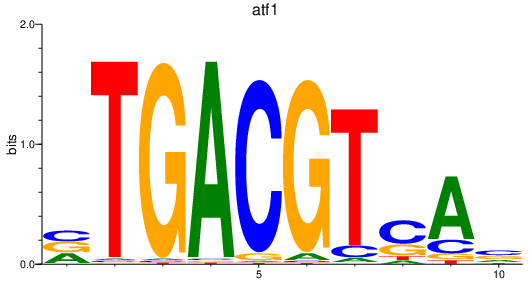
atf1
|
ENSDARG00000044301 | activating transcription factor 1 |
|
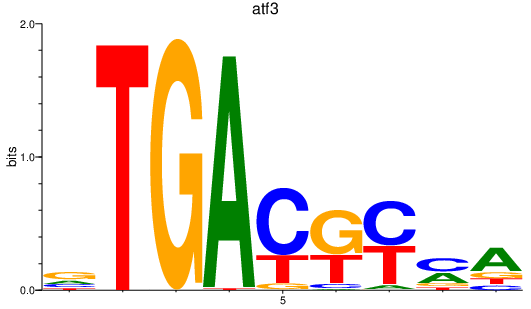
atf3
|
ENSDARG00000007823 | activating transcription factor 3 |
|
crema
|
ENSDARG00000023217 | cAMP responsive element modulator a |
|
cremb
|
ENSDARG00000102899 | cAMP responsive element modulator b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf3 | dr10_dc_chr20_+_37891739_37891747 | 0.88 | 5.7e-06 | Click! |
| crema | dr10_dc_chr2_-_43318809_43318849 | -0.87 | 1.1e-05 | Click! |
| atf1 | dr10_dc_chr6_-_39524075_39524296 | 0.85 | 3.4e-05 | Click! |
| atf2 | dr10_dc_chr9_+_2346341_2346400 | -0.58 | 1.7e-02 | Click! |
Activity profile of atf2_atf1_atf3_crema+cremb motif
Sorted Z-values of atf2_atf1_atf3_crema+cremb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of atf2_atf1_atf3_crema+cremb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_36512943 | 9.31 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr24_-_19573615 | 8.95 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr11_-_44539778 | 8.03 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr8_-_410190 | 8.00 |
ENSDART00000151155
|
trim36
|
tripartite motif containing 36 |
| chr7_+_17848688 | 7.45 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr17_+_25168837 | 7.40 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr25_+_34340569 | 7.16 |
ENSDART00000157638
|
tmem231
|
transmembrane protein 231 |
| chr2_+_35621136 | 7.02 |
ENSDART00000133018
ENSDART00000147278 |
plk3
|
polo-like kinase 3 (Drosophila) |
| chr24_-_19573966 | 6.90 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr19_+_14592632 | 6.85 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr2_-_57020663 | 6.78 |
|
|
|
| chr7_-_19989384 | 6.77 |
ENSDART00000173619
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr5_-_28931727 | 6.61 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr17_+_52526741 | 6.59 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr16_-_41585481 | 6.58 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr10_-_34971985 | 6.50 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr14_+_29945327 | 6.14 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr7_-_51497945 | 6.10 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr22_+_24596299 | 6.02 |
ENSDART00000158303
ENSDART00000160924 |
mcoln2
|
mucolipin 2 |
| chr4_-_16639774 | 6.00 |
ENSDART00000125323
|
caprin2
|
caprin family member 2 |
| chr16_-_42116471 | 5.94 |
ENSDART00000058620
|
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr8_+_52456064 | 5.77 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr22_+_1500412 | 5.76 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr15_+_5125138 | 5.68 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr10_+_7634671 | 5.66 |
ENSDART00000171744
|
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr23_-_1562572 | 5.42 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr17_-_24684870 | 5.41 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr23_+_20586745 | 5.38 |
ENSDART00000157522
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| KN150030v1_-_22572 | 5.37 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr2_-_31817448 | 5.35 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr20_-_20921480 | 5.31 |
ENSDART00000152781
|
ckbb
|
creatine kinase, brain b |
| chr22_+_1837102 | 5.24 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr12_-_24711074 | 4.99 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr10_+_15644868 | 4.91 |
ENSDART00000139259
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr24_+_12845339 | 4.89 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr8_-_410293 | 4.83 |
ENSDART00000164886
|
trim36
|
tripartite motif containing 36 |
| chr17_+_27988842 | 4.62 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr13_+_51485084 | 4.60 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr2_-_31817409 | 4.54 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr11_+_36117256 | 4.52 |
ENSDART00000141529
|
atxn7l2a
|
ataxin 7-like 2a |
| chr16_-_55210909 | 4.52 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr12_-_17741377 | 4.52 |
ENSDART00000166604
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr14_-_745312 | 4.52 |
ENSDART00000049918
ENSDART00000165856 |
trim35-27
|
tripartite motif containing 35-27 |
| chr2_+_9762781 | 4.46 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr19_+_14592246 | 4.43 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr15_+_5124690 | 4.42 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr16_-_5205600 | 4.39 |
ENSDART00000148955
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr6_+_94581 | 4.37 |
ENSDART00000125176
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr19_+_10742944 | 4.33 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr7_-_19989419 | 4.32 |
ENSDART00000127699
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr15_+_5124897 | 4.31 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr2_-_57020490 | 4.27 |
|
|
|
| chr18_-_5167807 | 4.27 |
|
|
|
| chr11_-_44859225 | 4.21 |
ENSDART00000163776
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr25_-_34340016 | 4.21 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr7_+_10319999 | 4.19 |
ENSDART00000168801
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_-_54391298 | 4.16 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr17_+_23534953 | 4.12 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr17_+_15527923 | 3.98 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr13_+_8508786 | 3.95 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr20_+_46668743 | 3.95 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr14_-_41101660 | 3.94 |
ENSDART00000003170
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr12_-_10474942 | 3.89 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr6_+_40994859 | 3.84 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr23_-_10850891 | 3.79 |
|
|
|
| chr15_+_37644266 | 3.71 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr9_-_27585644 | 3.70 |
ENSDART00000101401
|
tex30
|
testis expressed 30 |
| chr25_-_31352866 | 3.70 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr2_+_9762627 | 3.68 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr1_-_17000212 | 3.66 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr17_-_31195299 | 3.65 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr10_+_44456550 | 3.65 |
ENSDART00000157611
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr3_+_16772351 | 3.62 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr6_+_40994918 | 3.61 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr20_-_3301951 | 3.59 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr7_+_69211965 | 3.59 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr5_-_9035789 | 3.59 |
ENSDART00000124384
|
gak
|
cyclin G associated kinase |
| chr16_-_55211053 | 3.55 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr18_+_18890626 | 3.52 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr9_-_25555313 | 3.52 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| KN150030v1_-_22613 | 3.52 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr21_-_25991335 | 3.51 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr6_+_27676886 | 3.50 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr22_-_26971170 | 3.50 |
ENSDART00000162742
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr17_+_25163592 | 3.43 |
ENSDART00000100277
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr21_-_25991445 | 3.43 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr4_-_14916491 | 3.43 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr8_-_38284959 | 3.41 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| KN150455v1_-_14200 | 3.41 |
|
|
|
| chr22_+_1153361 | 3.41 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr5_+_56988663 | 3.41 |
ENSDART00000074268
|
zgc:153929
|
zgc:153929 |
| chr4_-_25526284 | 3.40 |
ENSDART00000142276
|
rbm17
|
RNA binding motif protein 17 |
| chr22_+_2422443 | 3.39 |
|
|
|
| chr16_-_34471672 | 3.38 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr2_-_37495925 | 3.38 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr7_+_1443102 | 3.34 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr8_+_23704663 | 3.33 |
ENSDART00000104346
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr19_-_12728868 | 3.33 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr23_-_25760917 | 3.32 |
|
|
|
| chr23_-_27579257 | 3.31 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr20_+_37891739 | 3.30 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr8_+_29733037 | 3.28 |
ENSDART00000133955
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr4_-_14916416 | 3.25 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr7_+_66410533 | 3.24 |
ENSDART00000027616
ENSDART00000162763 |
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr2_-_4191540 | 3.24 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr7_+_45747622 | 3.24 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr24_-_16835352 | 3.24 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr3_+_33608070 | 3.20 |
|
|
|
| chr10_-_34971926 | 3.19 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr16_-_29452509 | 3.19 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr23_+_10870124 | 3.19 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr13_-_49749441 | 3.19 |
ENSDART00000136165
|
lyst
|
lysosomal trafficking regulator |
| chr13_+_404116 | 3.17 |
ENSDART00000147909
|
wu:fc17b08
|
wu:fc17b08 |
| chr19_+_14592518 | 3.17 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr19_+_43090603 | 3.16 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr7_+_45747395 | 3.14 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr4_+_74723809 | 3.13 |
ENSDART00000172734
|
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr5_-_68011971 | 3.12 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr4_-_71752866 | 3.12 |
ENSDART00000143417
ENSDART00000172042 |
zgc:162958
|
zgc:162958 |
| chr6_+_40995061 | 3.11 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr3_-_16263615 | 3.10 |
ENSDART00000127309
|
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr25_-_6320310 | 3.10 |
ENSDART00000139606
|
snupn
|
snurportin 1 |
| chr13_-_36312975 | 3.08 |
ENSDART00000084867
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr21_-_25991595 | 3.08 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr17_+_15527861 | 3.07 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr2_-_56511961 | 3.06 |
ENSDART00000168160
|
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr12_-_22116694 | 3.06 |
ENSDART00000038310
ENSDART00000132731 |
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr6_+_59580554 | 3.06 |
|
|
|
| chr23_-_29952057 | 3.05 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr12_-_17741310 | 3.03 |
ENSDART00000166604
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr17_-_31195376 | 3.03 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr9_+_42744889 | 3.02 |
|
|
|
| chr17_+_23534824 | 3.02 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr12_-_926596 | 3.00 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr5_-_37655292 | 3.00 |
ENSDART00000156291
|
CT025690.1
|
ENSDARG00000096962 |
| chr10_+_32702525 | 2.99 |
ENSDART00000137244
|
zbtb21
|
zinc finger and BTB domain containing 21 |
| chr23_+_23256896 | 2.98 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr17_-_22552999 | 2.97 |
ENSDART00000141523
|
exo1
|
exonuclease 1 |
| chr3_-_18039574 | 2.97 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr22_+_1445227 | 2.96 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr6_+_19167599 | 2.94 |
ENSDART00000086619
|
prkca
|
protein kinase C, alpha |
| chr19_+_27338879 | 2.94 |
ENSDART00000149988
|
nelfe
|
negative elongation factor complex member E |
| chr2_-_6380292 | 2.94 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr6_-_54436651 | 2.94 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr22_-_760140 | 2.93 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr3_-_45358502 | 2.92 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr5_+_61711614 | 2.90 |
ENSDART00000082965
|
ABR
|
active BCR-related |
| chr23_+_33792145 | 2.90 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr15_+_5124616 | 2.88 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr12_-_10384031 | 2.83 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr18_+_6417959 | 2.82 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr17_+_15527525 | 2.82 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr21_+_3624059 | 2.82 |
ENSDART00000111699
|
tor1
|
torsin family 1 |
| chr16_-_41585555 | 2.82 |
ENSDART00000131706
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr21_+_3764216 | 2.79 |
ENSDART00000149777
|
setx
|
senataxin |
| chr7_+_10320142 | 2.79 |
ENSDART00000168801
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr25_-_22954929 | 2.78 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr10_+_14530451 | 2.78 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr5_+_12971972 | 2.78 |
ENSDART00000132406
|
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr12_-_24711169 | 2.78 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr3_-_30778382 | 2.78 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr7_+_28341426 | 2.78 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr5_+_71360249 | 2.78 |
|
|
|
| chr19_+_14592757 | 2.77 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr8_-_38284748 | 2.77 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr12_+_33793663 | 2.77 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr2_+_22753634 | 2.76 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr15_-_41732606 | 2.76 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr10_+_27028355 | 2.75 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr6_-_10677028 | 2.75 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr5_-_9036049 | 2.73 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr4_+_9010972 | 2.73 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr1_+_9621488 | 2.73 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr10_-_2944190 | 2.72 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr17_-_7635061 | 2.72 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr11_-_13283709 | 2.71 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| KN150699v1_-_15078 | 2.71 |
ENSDART00000159861
|
ENSDARG00000098739
|
ENSDARG00000098739 |
| chr10_-_6587281 | 2.69 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr14_+_31278584 | 2.69 |
ENSDART00000158875
ENSDART00000026195 |
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr17_+_27988973 | 2.68 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr25_-_36513113 | 2.67 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr15_-_29229170 | 2.67 |
ENSDART00000138449
|
xaf1
|
XIAP associated factor 1 |
| chr7_+_37818298 | 2.67 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr18_-_16964811 | 2.65 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr11_-_22663375 | 2.65 |
ENSDART00000154813
|
mdm4
|
MDM4, p53 regulator |
| chr18_+_22804127 | 2.64 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr23_+_17583383 | 2.64 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr15_-_778461 | 2.64 |
ENSDART00000153874
|
si:dkey-7i4.19
|
si:dkey-7i4.19 |
| chr23_+_23256859 | 2.64 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr8_-_38284904 | 2.64 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_+_3167367 | 2.62 |
ENSDART00000160604
|
ftsj3
|
FtsJ homolog 3 (E. coli) |
| chr19_-_3547196 | 2.62 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr3_-_40791035 | 2.60 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr20_-_3301898 | 2.59 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr20_-_44678938 | 2.59 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr8_-_16638825 | 2.58 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr12_+_46853062 | 2.56 |
ENSDART00000171146
|
hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr22_-_10861268 | 2.55 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr22_+_1451958 | 2.55 |
|
|
|
| chr24_+_16838877 | 2.55 |
ENSDART00000145520
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:1900144 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 3.0 | 12.0 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 2.3 | 7.0 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 2.2 | 8.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 1.6 | 7.8 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 1.5 | 12.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 1.5 | 6.1 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 1.5 | 6.0 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 1.5 | 16.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 1.5 | 5.8 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 1.5 | 2.9 | GO:0071331 | cellular glucose homeostasis(GO:0001678) cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 1.3 | 9.2 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 1.3 | 3.9 | GO:0072540 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) type 2 immune response(GO:0042092) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T cell selection(GO:0045058) T-helper 2 cell differentiation(GO:0045064) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 1.2 | 9.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.2 | 9.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 1.1 | 4.5 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 1.0 | 4.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 1.0 | 3.1 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 1.0 | 3.1 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 1.0 | 3.1 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 1.0 | 6.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 1.0 | 2.9 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 1.0 | 2.9 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.9 | 7.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.9 | 2.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.9 | 3.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.9 | 3.7 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.9 | 4.4 | GO:0032516 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.9 | 2.7 | GO:0006408 | snRNA export from nucleus(GO:0006408) regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) snRNA transport(GO:0051030) |
| 0.9 | 4.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.9 | 2.6 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.8 | 0.8 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.8 | 0.8 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.8 | 6.4 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.8 | 2.4 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.7 | 23.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.7 | 2.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.7 | 4.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.7 | 9.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.6 | 8.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 1.9 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.6 | 2.5 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.6 | 3.1 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.6 | 2.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.6 | 2.3 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of purine nucleotide metabolic process(GO:1900543) |
| 0.6 | 2.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.6 | 4.0 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.6 | 2.8 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.6 | 5.0 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.5 | 8.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.5 | 5.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.5 | 10.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.5 | 3.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 5.7 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.5 | 2.6 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.5 | 3.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.5 | 2.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.5 | 4.0 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.5 | 2.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.5 | 1.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 2.4 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.5 | 0.5 | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.5 | 2.9 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 2.9 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.5 | 3.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.5 | 2.8 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 1.9 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.5 | 1.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.5 | 3.7 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.5 | 3.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 1.8 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.4 | 4.0 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.4 | 3.6 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.4 | 1.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.4 | 2.5 | GO:0048608 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.4 | 8.0 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.4 | 0.4 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.4 | 5.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.4 | 0.8 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.4 | 6.2 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.4 | 2.4 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.4 | 12.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.4 | 3.6 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.4 | 1.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 1.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.4 | 2.7 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.4 | 1.9 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.4 | 2.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 1.8 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 2.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 5.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 3.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 2.8 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.3 | 0.7 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.3 | 1.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.3 | 6.9 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.3 | 8.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 4.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.3 | 1.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 1.3 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.3 | 1.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.3 | 0.9 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.3 | 1.8 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.3 | 2.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 3.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.3 | 3.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 2.0 | GO:0055015 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.3 | 5.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 10.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.3 | 2.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 0.8 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.3 | 1.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.3 | 1.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 12.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.3 | 0.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 4.3 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.3 | 0.8 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 0.7 | GO:0072501 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 2.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.2 | 3.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 12.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 0.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 11.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 4.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.2 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 10.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 0.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 2.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 5.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 3.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 1.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 1.6 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.2 | 2.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.8 | GO:1990379 | lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.2 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 1.4 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.2 | 1.4 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.2 | 1.0 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.2 | 1.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.7 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.2 | 1.7 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 0.8 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 2.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 0.7 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.7 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.2 | 1.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 5.8 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.2 | 1.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 0.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 2.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 1.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 2.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 3.8 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 2.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 4.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 3.9 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.2 | 1.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 0.9 | GO:0051589 | negative regulation of synaptic transmission(GO:0050805) negative regulation of neurotransmitter transport(GO:0051589) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.4 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 1.7 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 5.9 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 1.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 8.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.7 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.9 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.1 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.2 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.1 | 0.2 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.4 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.1 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 1.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 5.9 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 1.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 6.1 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.1 | 0.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 3.3 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.1 | 0.1 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 1.2 | GO:0051351 | regulation of ligase activity(GO:0051340) positive regulation of ligase activity(GO:0051351) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.7 | GO:0060232 | delamination(GO:0060232) |
| 0.1 | 1.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.3 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 3.5 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 0.7 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 2.2 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 2.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.3 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.1 | 2.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.8 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.1 | 4.8 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 4.0 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) mitochondrial protein processing(GO:0034982) |
| 0.1 | 1.0 | GO:0009070 | serine family amino acid biosynthetic process(GO:0009070) |
| 0.1 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.6 | GO:0030534 | adult behavior(GO:0030534) |
| 0.1 | 1.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 2.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.2 | GO:1903003 | regulation of protein deubiquitination(GO:0090085) positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.7 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 1.8 | GO:0009994 | oocyte differentiation(GO:0009994) oocyte development(GO:0048599) |
| 0.1 | 0.6 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 1.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 2.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.9 | GO:0071428 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 1.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.2 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.2 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.1 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.0 | GO:0043588 | skin development(GO:0043588) |
| 0.1 | 6.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.7 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.2 | GO:2000104 | replication fork protection(GO:0048478) negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.1 | 3.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.9 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.6 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.1 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 0.6 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.6 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.8 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.4 | GO:1902743 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.0 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 6.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) ornithine metabolic process(GO:0006591) urea metabolic process(GO:0019627) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 18.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.0 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.6 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 1.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 3.0 | GO:0045216 | cell-cell junction organization(GO:0045216) |
| 0.0 | 1.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:0051792 | response to progesterone(GO:0032570) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.3 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.0 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 7.2 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 3.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 1.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 3.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 1.0 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.6 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 1.6 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 1.1 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.8 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 1.6 | 4.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.4 | 9.9 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.3 | 5.3 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 1.3 | 9.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 1.3 | 3.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.1 | 6.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 1.1 | 5.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.0 | 3.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.0 | 3.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.9 | 4.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.8 | 6.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.7 | 2.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.7 | 3.5 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.7 | 3.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.7 | 2.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.6 | 1.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.6 | 1.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.6 | 11.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 3.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.6 | 3.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.6 | 1.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.5 | 5.9 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.5 | 2.7 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.5 | 3.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.4 | 3.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 4.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.4 | 22.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.4 | 1.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.4 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.4 | 10.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 8.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.3 | 1.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 2.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 11.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 1.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 0.9 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 2.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 1.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 3.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 0.8 | GO:1990077 | primosome complex(GO:1990077) |
| 0.3 | 8.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 2.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 1.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 0.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 2.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 17.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 1.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 0.4 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.2 | 1.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 2.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 3.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 0.9 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 3.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 2.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 2.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 4.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 4.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 4.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 3.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 6.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.5 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 7.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 1.8 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 10.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 5.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 7.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.8 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 2.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 7.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 17.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 1.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 2.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 7.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 4.8 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 7.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 3.7 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.8 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 15.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 2.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 12.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 10.2 | GO:0031966 | mitochondrial envelope(GO:0005740) mitochondrial membrane(GO:0031966) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.0 | 0.2 | GO:0070993 | eukaryotic 43S preinitiation complex(GO:0016282) translation preinitiation complex(GO:0070993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 1.5 | 10.8 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 1.5 | 7.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 1.5 | 5.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 1.3 | 5.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 1.2 | 7.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 1.2 | 3.6 | GO:0017125 | deoxycytidyl transferase activity(GO:0017125) |
| 1.1 | 13.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.1 | 4.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 1.0 | 8.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 1.0 | 18.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.9 | 5.6 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.9 | 3.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.9 | 12.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.8 | 8.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.8 | 6.4 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.8 | 2.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.8 | 2.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.8 | 3.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 4.3 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.7 | 2.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.6 | 23.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 2.3 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.6 | 3.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 2.2 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.5 | 7.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.5 | 7.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.5 | 2.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.5 | 1.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 1.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 6.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.5 | 2.8 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 2.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.5 | 2.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.4 | 3.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 2.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 1.3 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 1.3 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.4 | 5.0 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.4 | 2.9 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.4 | 8.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.4 | 2.4 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.4 | 1.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.4 | 13.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.4 | 2.6 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.3 | 5.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 2.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 2.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 2.0 | GO:1901611 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.3 | 1.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.3 | 1.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 1.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 5.3 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.3 | 1.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 2.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.3 | 9.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 1.4 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.3 | 1.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.3 | 25.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.3 | 3.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.3 | 1.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 1.0 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.3 | 3.4 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.3 | 0.8 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 8.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 1.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 2.4 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 16.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.2 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 0.9 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 4.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 9.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.2 | 1.8 | GO:0004322 | ferroxidase activity(GO:0004322) ferric iron binding(GO:0008199) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 15.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 4.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 0.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 1.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 6.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.8 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.2 | 2.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.4 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 3.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 8.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 1.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 1.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 13.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.2 | 0.9 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 1.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 3.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 0.5 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 3.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 14.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.7 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 9.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.9 | GO:0030291 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.1 | 0.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.1 | 1.1 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 2.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.3 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 1.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 3.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 5.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 31.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 7.2 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 6.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.3 | GO:0043878 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 3.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 4.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.0 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 3.7 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 2.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.6 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 5.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 1.2 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 4.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 11.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.9 | 14.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.5 | 2.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 3.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 11.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 7.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 11.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 0.7 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 2.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 9.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 4.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 1.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 7.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 8.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.9 | 5.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.8 | 4.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.8 | 5.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.8 | 1.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.7 | 15.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.5 | 10.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 8.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 3.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 2.8 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.4 | 4.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 2.0 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 2.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 3.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 2.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 5.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.3 | 4.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 9.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 1.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 6.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 0.6 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 0.2 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.2 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 3.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 4.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 7.1 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 3.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 3.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 5.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 22.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.0 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 0.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 4.0 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.1 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 4.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |