Project
DANIO-CODE
Navigation
Downloads
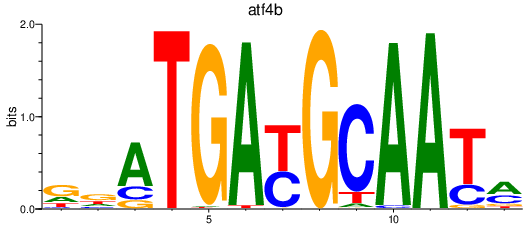
Results for atf4b
Z-value: 0.96
Transcription factors associated with atf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf4b
|
ENSDARG00000038141 | activating transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf4b | dr10_dc_chr3_+_24067430_24067435 | -0.86 | 2.0e-05 | Click! |
Activity profile of atf4b motif
Sorted Z-values of atf4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of atf4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_37891739 | 4.62 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr20_+_37891795 | 3.55 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr12_+_30673985 | 2.87 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr14_-_7437682 | 2.43 |
ENSDART00000172215
|
si:ch211-39i2.2
|
si:ch211-39i2.2 |
| chr12_+_17032829 | 2.33 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr18_+_17611571 | 2.23 |
ENSDART00000151850
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr19_+_43745687 | 2.22 |
ENSDART00000102384
|
sesn2
|
sestrin 2 |
| chr19_+_43745546 | 2.22 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr18_+_17611931 | 2.10 |
ENSDART00000141465
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr12_+_17032997 | 2.05 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr18_+_17611865 | 2.01 |
ENSDART00000141465
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr3_-_40142513 | 1.96 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr19_+_43745515 | 1.74 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr19_-_8893920 | 1.65 |
ENSDART00000025385
|
cers2a
|
ceramide synthase 2a |
| chr21_+_30684976 | 1.62 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr14_+_7585289 | 1.58 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr20_+_28367799 | 1.50 |
ENSDART00000153319
ENSDART00000103330 |
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr3_+_40142679 | 1.49 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr24_+_16838877 | 1.47 |
ENSDART00000145520
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr5_+_56988663 | 1.45 |
ENSDART00000074268
|
zgc:153929
|
zgc:153929 |
| chr12_+_17032963 | 1.44 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr3_+_15244806 | 1.44 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr7_+_71263100 | 1.39 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr12_-_46943717 | 1.34 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr9_-_48516764 | 1.34 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr23_+_25062826 | 1.30 |
ENSDART00000172299
|
arhgap4a
|
Rho GTPase activating protein 4a |
| chr7_+_71262845 | 1.21 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr9_+_2523927 | 1.15 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr3_+_25776714 | 1.15 |
ENSDART00000170324
|
tom1
|
target of myb1 membrane trafficking protein |
| chr10_-_8839096 | 1.13 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr20_-_20711298 | 1.13 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr18_+_17611519 | 1.11 |
ENSDART00000175258
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr9_+_10728013 | 1.04 |
ENSDART00000130065
|
dars
|
aspartyl-tRNA synthetase |
| chr3_+_33308831 | 1.03 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr5_-_34600655 | 1.02 |
ENSDART00000164396
|
fcho2
|
FCH domain only 2 |
| chr17_-_42523585 | 1.01 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr7_+_15781933 | 1.01 |
ENSDART00000161669
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr24_+_16838984 | 1.00 |
ENSDART00000023833
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr12_+_13167278 | 1.00 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr3_-_30022268 | 0.99 |
ENSDART00000159065
|
nucb1
|
nucleobindin 1 |
| chr21_+_30685020 | 0.99 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr14_-_7585265 | 0.96 |
|
|
|
| chr18_-_18678313 | 0.94 |
ENSDART00000076695
|
aars
|
alanyl-tRNA synthetase |
| chr17_-_42523635 | 0.93 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr12_+_13167781 | 0.90 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr14_+_28132910 | 0.89 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long-chain family member 4a |
| chr9_-_24407859 | 0.87 |
ENSDART00000135356
|
nabp1a
|
nucleic acid binding protein 1a |
| chr14_-_30604897 | 0.86 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr10_+_16914003 | 0.82 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr15_+_24802510 | 0.82 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr4_+_16795681 | 0.79 |
ENSDART00000134054
|
si:dkey-13i19.8
|
si:dkey-13i19.8 |
| chr23_-_26301505 | 0.77 |
ENSDART00000162423
|
ENSDARG00000098968
|
ENSDARG00000098968 |
| chr5_-_29551051 | 0.77 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr22_+_25911875 | 0.76 |
ENSDART00000157842
|
dnaja3b
|
DnaJ (Hsp40) homolog, subfamily A, member 3B |
| chr3_-_40142617 | 0.76 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr18_+_20058174 | 0.73 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr3_-_40142333 | 0.71 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr6_-_4067642 | 0.71 |
ENSDART00000162497
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr4_+_16795573 | 0.70 |
ENSDART00000134054
|
si:dkey-13i19.8
|
si:dkey-13i19.8 |
| chr23_-_43916621 | 0.67 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr12_-_13167362 | 0.67 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr4_+_16795523 | 0.66 |
ENSDART00000134054
|
si:dkey-13i19.8
|
si:dkey-13i19.8 |
| chr12_-_48391342 | 0.66 |
ENSDART00000153209
|
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr8_-_31375316 | 0.65 |
ENSDART00000159878
ENSDART00000164134 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr19_+_31297635 | 0.65 |
ENSDART00000139599
|
yars
|
tyrosyl-tRNA synthetase |
| chr6_+_49552893 | 0.63 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr9_-_48516909 | 0.63 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr14_+_7585533 | 0.63 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr14_-_23504183 | 0.63 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_-_7000549 | 0.62 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr5_-_68751287 | 0.61 |
ENSDART00000112692
|
ENSDARG00000077155
|
ENSDARG00000077155 |
| chr23_+_43916520 | 0.59 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr8_+_42910733 | 0.59 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr20_-_20711525 | 0.58 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr8_+_22305999 | 0.57 |
ENSDART00000075129
|
lrrc47
|
leucine rich repeat containing 47 |
| chr3_-_30022354 | 0.56 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr23_-_29467861 | 0.53 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr16_-_42866318 | 0.49 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr6_+_23709605 | 0.46 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr23_+_43916552 | 0.46 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr23_+_38313746 | 0.44 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr16_-_42866236 | 0.42 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr20_-_19632944 | 0.42 |
ENSDART00000125830
|
eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr8_+_7739964 | 0.40 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr7_+_28814408 | 0.40 |
ENSDART00000173960
|
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr14_+_28860508 | 0.38 |
ENSDART00000166608
|
ENSDARG00000037122
|
ENSDARG00000037122 |
| chr11_-_23826046 | 0.38 |
|
|
|
| chr12_+_34851952 | 0.37 |
ENSDART00000123988
|
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr4_+_25618251 | 0.36 |
ENSDART00000044739
ENSDART00000028297 |
acot15
acot14
|
acyl-CoA thioesterase 15 acyl-CoA thioesterase 14 |
| chr15_+_24802607 | 0.34 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr11_+_44909949 | 0.29 |
ENSDART00000173116
ENSDART00000167011 ENSDART00000167226 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr21_+_9904707 | 0.28 |
ENSDART00000171579
|
herc7
|
hect domain and RLD 7 |
| chr15_-_7340623 | 0.28 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
solute carrier family 7 member 1 |
| chr24_-_38757086 | 0.27 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr16_-_13927947 | 0.26 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr13_-_8968136 | 0.25 |
ENSDART00000144714
|
HTRA2 (1 of many)
|
HtrA serine peptidase 2 |
| chr20_+_28901045 | 0.24 |
ENSDART00000153046
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr24_+_32610480 | 0.24 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr16_-_13927684 | 0.23 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr15_+_24802238 | 0.21 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr15_+_24802704 | 0.19 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr9_-_48516792 | 0.19 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr8_+_7740024 | 0.18 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr5_-_34600498 | 0.18 |
ENSDART00000164396
|
fcho2
|
FCH domain only 2 |
| chr21_+_3733019 | 0.12 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr6_-_9471808 | 0.11 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr2_+_35258438 | 0.10 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr14_-_23503974 | 0.08 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr7_+_71262935 | 0.08 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr15_+_20607816 | 0.06 |
ENSDART00000169941
|
sgsm2
|
small G protein signaling modulator 2 |
| chr3_-_15739926 | 0.06 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr5_-_68727127 | 0.05 |
ENSDART00000023983
|
psat1
|
phosphoserine aminotransferase 1 |
| chr25_-_5921772 | 0.05 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr17_+_8018808 | 0.04 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr6_-_58921392 | 0.03 |
|
|
|
| chr14_+_37203442 | 0.03 |
|
|
|
| chr25_+_28450465 | 0.03 |
ENSDART00000075151
|
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr13_-_8968218 | 0.03 |
ENSDART00000144142
ENSDART00000144714 |
HTRA2 (1 of many)
|
HtrA serine peptidase 2 |
| chr5_+_43365528 | 0.02 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr23_-_43916434 | 0.01 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr3_+_33309011 | 0.01 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.9 | 7.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.6 | 2.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.5 | 2.7 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.4 | 2.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 2.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 1.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 1.8 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.3 | 1.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.7 | GO:0071387 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.2 | 0.9 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.2 | 8.2 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 0.6 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.4 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.7 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.1 | 0.5 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.1 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.9 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 0.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 1.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.6 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.0 | 0.7 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 2.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 2.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 10.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.3 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.7 | 2.2 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.3 | 7.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 1.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.9 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 2.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 1.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 13.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.3 | 3.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.7 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |