Project
DANIO-CODE
Navigation
Downloads
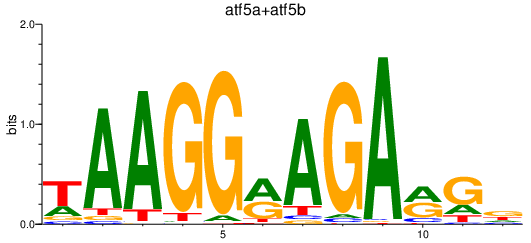
Results for atf5a+atf5b
Z-value: 2.10
Transcription factors associated with atf5a+atf5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf5a
|
ENSDARG00000068096 | activating transcription factor 5a |
|
atf5b
|
ENSDARG00000077785 | activating transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf5b | dr10_dc_chr15_-_17933972_17934075 | 0.94 | 7.0e-08 | Click! |
| atf5a | dr10_dc_chr5_-_29818325_29818328 | -0.74 | 1.1e-03 | Click! |
Activity profile of atf5a+atf5b motif
Sorted Z-values of atf5a+atf5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of atf5a+atf5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_5048203 | 5.95 |
ENSDART00000150863
ENSDART00000163417 |
nrp2a
|
neuropilin 2a |
| chr22_-_13139894 | 5.60 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr11_-_29410649 | 5.50 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr16_-_24602919 | 5.47 |
ENSDART00000147478
|
cadm4
|
cell adhesion molecule 4 |
| chr3_-_48865474 | 5.43 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr9_+_25964943 | 5.21 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr11_+_5929544 | 5.12 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr16_-_24220413 | 4.79 |
ENSDART00000103176
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr7_+_35769973 | 4.71 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| chr7_+_60054116 | 4.69 |
ENSDART00000145201
|
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr3_+_36146072 | 4.66 |
ENSDART00000148444
|
zgc:86896
|
zgc:86896 |
| chr24_+_9231693 | 4.62 |
ENSDART00000082422
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr13_+_23065500 | 4.48 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_-_19646761 | 4.46 |
|
|
|
| chr5_+_37053530 | 4.38 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr5_+_38487183 | 4.17 |
ENSDART00000164065
|
anxa3a
|
annexin A3a |
| chr7_-_37816600 | 4.03 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr4_+_7668939 | 3.95 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr7_+_25052543 | 3.90 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr25_-_32344881 | 3.78 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr4_-_28346021 | 3.64 |
ENSDART00000178149
|
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr20_-_26521700 | 3.60 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr13_-_22877318 | 3.58 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr15_+_37204488 | 3.52 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr11_-_23151247 | 3.49 |
|
|
|
| chr5_-_2468736 | 3.47 |
ENSDART00000170112
|
rabepk
|
Rab9 effector protein with kelch motifs |
| chr18_-_23889256 | 3.42 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_30932612 | 3.37 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr20_+_30894667 | 3.31 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr14_+_13919450 | 3.30 |
ENSDART00000174760
|
nlgn3a
|
neuroligin 3a |
| chr17_+_38528709 | 3.18 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr5_-_47656197 | 3.08 |
|
|
|
| chr5_-_67233396 | 3.08 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr8_+_22268070 | 2.97 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr10_-_7427767 | 2.88 |
ENSDART00000167963
ENSDART00000162021 |
nrg1
|
neuregulin 1 |
| chr19_+_20209561 | 2.87 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr1_+_40795366 | 2.84 |
ENSDART00000146310
|
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr9_+_25964868 | 2.82 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr12_-_3742062 | 2.77 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr14_+_5078937 | 2.74 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr5_-_1698166 | 2.70 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr22_+_36687668 | 2.66 |
ENSDART00000134031
|
b3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr16_-_16796313 | 2.65 |
|
|
|
| chr3_+_13692658 | 2.65 |
ENSDART00000162317
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr7_+_60054196 | 2.62 |
ENSDART00000039827
|
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr23_-_34970838 | 2.61 |
ENSDART00000087219
|
ENSDARG00000061272
|
ENSDARG00000061272 |
| chr24_-_16993914 | 2.60 |
|
|
|
| chr3_+_23573114 | 2.54 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr13_+_30743690 | 2.51 |
ENSDART00000134809
|
ercc6
|
excision repair cross-complementation group 6 |
| chr12_-_47699958 | 2.48 |
ENSDART00000171932
ENSDART00000168165 ENSDART00000161985 |
HHEX
|
hematopoietically expressed homeobox |
| chr5_+_18740715 | 2.46 |
ENSDART00000089173
|
atp8b5a
|
ATPase, class I, type 8B, member 5a |
| chr9_-_35260503 | 2.46 |
ENSDART00000125536
|
appb
|
amyloid beta (A4) precursor protein b |
| chr17_-_37107310 | 2.44 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr16_+_17705704 | 2.36 |
|
|
|
| chr15_-_27431216 | 2.33 |
ENSDART00000155933
|
AL954322.2
|
ENSDARG00000097072 |
| chr10_+_4874841 | 2.30 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr16_+_10531986 | 2.28 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr23_-_28390747 | 2.24 |
|
|
|
| chr5_-_63531729 | 2.23 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr10_-_7827244 | 2.21 |
ENSDART00000111058
|
mpx
|
myeloid-specific peroxidase |
| chr3_+_28450576 | 2.19 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr18_-_19467100 | 2.16 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr3_+_37565550 | 2.15 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr18_-_23888988 | 2.15 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr13_+_30097568 | 2.13 |
ENSDART00000134691
|
rps24
|
ribosomal protein S24 |
| chr8_-_37011436 | 2.10 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr24_+_9231768 | 2.10 |
ENSDART00000082422
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr18_-_23889025 | 2.08 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_28303542 | 2.07 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr3_-_16077831 | 2.07 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr12_-_3741925 | 2.07 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr15_-_19314712 | 2.03 |
ENSDART00000092705
|
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr18_-_50156084 | 2.00 |
ENSDART00000006078
|
loxl1
|
lysyl oxidase-like 1 |
| chr4_-_16344954 | 1.97 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr22_-_28840467 | 1.95 |
|
|
|
| chr12_-_7902815 | 1.95 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr10_+_35344246 | 1.93 |
ENSDART00000139229
|
pora
|
P450 (cytochrome) oxidoreductase a |
| chr20_+_6640497 | 1.91 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr23_-_30114528 | 1.85 |
ENSDART00000131209
|
ccdc187
|
coiled-coil domain containing 187 |
| chr7_+_25052687 | 1.82 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr12_+_20578266 | 1.80 |
ENSDART00000016099
|
ENSDARG00000043818
|
ENSDARG00000043818 |
| chr24_+_24924379 | 1.79 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr21_-_26655339 | 1.79 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_+_10531926 | 1.74 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr7_-_13118705 | 1.71 |
ENSDART00000081212
|
polr2l
|
polymerase (RNA) II (DNA directed) polypeptide L |
| chr6_-_34024063 | 1.66 |
ENSDART00000003701
|
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr18_+_22804322 | 1.64 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr19_+_30212276 | 1.62 |
|
|
|
| chr18_-_23889169 | 1.62 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_-_46649324 | 1.58 |
|
|
|
| chr22_+_1568413 | 1.53 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr24_+_40292617 | 1.53 |
|
|
|
| chr3_-_48865399 | 1.50 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_17564926 | 1.48 |
|
|
|
| chr11_+_26238579 | 1.47 |
ENSDART00000165097
|
unm_sa1614
|
un-named sa1614 |
| chr21_-_22078508 | 1.46 |
ENSDART00000122389
|
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr4_+_7668691 | 1.46 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr1_+_31788994 | 1.43 |
ENSDART00000152580
|
nlgn4a
|
neuroligin 4a |
| chr17_-_37266525 | 1.41 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr16_-_6318609 | 1.39 |
|
|
|
| chr8_-_50299273 | 1.38 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr1_+_46537333 | 1.37 |
|
|
|
| chr8_+_22559432 | 1.34 |
|
|
|
| chr7_-_29956705 | 1.29 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr20_-_31524559 | 1.29 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr11_+_18020191 | 1.25 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr14_-_46649039 | 1.24 |
|
|
|
| chr1_-_42078442 | 1.23 |
ENSDART00000074618
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr18_+_22228682 | 1.21 |
ENSDART00000165464
|
fam65a
|
family with sequence similarity 65, member A |
| chr13_-_5423165 | 1.20 |
|
|
|
| chr4_-_16344835 | 1.19 |
ENSDART00000134449
|
epyc
|
epiphycan |
| chr16_+_17706003 | 1.19 |
|
|
|
| chr3_+_26015378 | 1.17 |
|
|
|
| chr5_-_25586926 | 1.17 |
ENSDART00000137027
|
ccdc125
|
coiled-coil domain containing 125 |
| chr1_+_23093475 | 1.15 |
|
|
|
| chr19_-_11161797 | 1.15 |
|
|
|
| chr14_-_46649231 | 1.14 |
|
|
|
| chr19_-_36009321 | 1.12 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr5_-_25586964 | 1.11 |
ENSDART00000163601
|
ccdc125
|
coiled-coil domain containing 125 |
| chr10_-_20524670 | 1.09 |
|
|
|
| chr15_-_40115930 | 1.06 |
ENSDART00000157388
|
lypc
|
ly6 domain containing, pigment cell |
| chr9_-_33975936 | 1.06 |
ENSDART00000140779
|
fundc1
|
FUN14 domain containing 1 |
| chr12_-_3742020 | 1.04 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr2_-_59013059 | 1.03 |
|
|
|
| chr24_-_20176227 | 1.02 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr16_-_29599005 | 1.01 |
|
|
|
| chr20_+_44684239 | 0.99 |
ENSDART00000149775
|
atad2b
|
ATPase family, AAA domain containing 2B |
| chr17_-_21773194 | 0.98 |
ENSDART00000160296
|
hmx3a
|
H6 family homeobox 3a |
| chr5_-_64637049 | 0.94 |
ENSDART00000164649
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr23_+_36648211 | 0.93 |
|
|
|
| chr17_-_20830287 | 0.93 |
ENSDART00000149630
|
ank3a
|
ankyrin 3a |
| chr8_-_17692185 | 0.92 |
|
|
|
| chr18_+_17796754 | 0.90 |
|
|
|
| chr9_+_30909382 | 0.87 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr9_+_30909291 | 0.84 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr16_+_11027394 | 0.80 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr3_-_32834113 | 0.77 |
ENSDART00000125012
|
casp6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr7_+_35770134 | 0.74 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| chr18_+_29010470 | 0.66 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr10_-_20524586 | 0.65 |
|
|
|
| chr11_+_16018035 | 0.63 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr9_+_6609374 | 0.60 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr15_+_42548643 | 0.58 |
|
|
|
| chr13_-_34926382 | 0.58 |
ENSDART00000178470
|
btbd3b
|
BTB (POZ) domain containing 3b |
| chr17_-_29885237 | 0.57 |
ENSDART00000009104
|
esrrga
|
estrogen-related receptor gamma a |
| chr12_+_29140448 | 0.55 |
|
|
|
| chr7_+_39353697 | 0.55 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr14_+_26424717 | 0.54 |
|
|
|
| chr11_-_25086785 | 0.50 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr7_+_39181175 | 0.49 |
|
|
|
| chr19_+_5054263 | 0.43 |
|
|
|
| chr24_+_34720673 | 0.41 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr13_-_44708149 | 0.41 |
|
|
|
| chr10_+_4874800 | 0.41 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr8_-_23239664 | 0.40 |
|
|
|
| chr20_-_31524614 | 0.40 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr10_-_45134642 | 0.40 |
ENSDART00000166528
|
purbb
|
purine-rich element binding protein Bb |
| chr20_-_26517630 | 0.36 |
|
|
|
| chr5_+_63716392 | 0.30 |
ENSDART00000015940
|
edf1
|
endothelial differentiation-related factor 1 |
| chr12_+_18622682 | 0.29 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr20_+_27341026 | 0.25 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr4_-_70948610 | 0.24 |
ENSDART00000174299
|
CABZ01054394.2
|
ENSDARG00000098324 |
| chr8_+_22268039 | 0.23 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr19_-_5142538 | 0.23 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr4_-_13256905 | 0.23 |
ENSDART00000026593
ENSDART00000150577 |
grip1
|
glutamate receptor interacting protein 1 |
| chr5_-_3077874 | 0.20 |
|
|
|
| chr3_+_19186724 | 0.18 |
ENSDART00000110548
|
kri1
|
KRI1 homolog |
| chr10_-_20524860 | 0.15 |
|
|
|
| chr8_-_16614882 | 0.12 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr13_-_743751 | 0.12 |
ENSDART00000167256
|
ENSDARG00000102651
|
ENSDARG00000102651 |
| chr15_-_8786861 | 0.12 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr3_+_23573053 | 0.10 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr3_-_26211047 | 0.10 |
ENSDART00000087196
|
zgc:153240
|
zgc:153240 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 1.9 | 9.3 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.4 | 5.6 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 1.3 | 8.0 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 1.1 | 3.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 1.0 | 2.9 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.9 | 2.7 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.9 | 5.5 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.8 | 3.8 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.7 | 5.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.4 | 5.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.4 | 4.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.4 | 2.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 2.0 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) protein oxidation(GO:0018158) |
| 0.3 | 3.6 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.3 | 6.0 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.3 | 1.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 3.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 2.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 1.4 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.2 | 4.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 2.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 2.2 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.2 | 1.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 2.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 4.4 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 4.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 1.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.8 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 1.7 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.1 | 2.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 5.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 2.4 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 6.6 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 0.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 3.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 2.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 3.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 2.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 4.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 2.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 2.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 6.9 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 1.0 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 1.1 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 3.0 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.6 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 1.9 | GO:0009725 | response to hormone(GO:0009725) |
| 0.0 | 1.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 3.4 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 5.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 4.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 4.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 4.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.2 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 4.0 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 7.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 6.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.7 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.8 | 9.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.7 | 6.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 3.6 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 2.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 2.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.5 | 4.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.5 | 6.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 5.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.4 | 1.9 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.4 | 3.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 2.6 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 3.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 6.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 4.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 6.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 13.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 2.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 8.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 26.7 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 10.2 | GO:0030234 | enzyme regulator activity(GO:0030234) |
| 0.0 | 7.0 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 1.6 | GO:0042802 | identical protein binding(GO:0042802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 7.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 2.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 2.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 5.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 4.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 2.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 16.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 9.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 4.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |