Project
DANIO-CODE
Navigation
Downloads
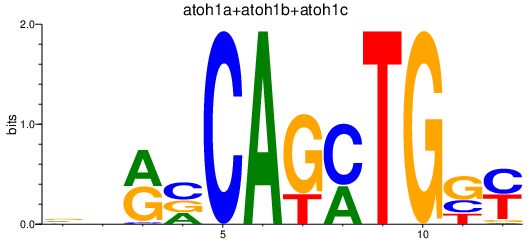
Results for atoh1a+atoh1b+atoh1c
Z-value: 0.81
Transcription factors associated with atoh1a+atoh1b+atoh1c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atoh1c
|
ENSDARG00000039602 | atonal bHLH transcription factor 1c |
|
atoh1a
|
ENSDARG00000055294 | atonal bHLH transcription factor 1a |
|
atoh1b
|
ENSDARG00000099564 | atonal bHLH transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atoh1a | dr10_dc_chr8_+_29584750_29584757 | 0.45 | 8.2e-02 | Click! |
| atoh1b | dr10_dc_chrUn_KN150642v1_+_9419_9454 | 0.29 | 2.8e-01 | Click! |
Activity profile of atoh1a+atoh1b+atoh1c motif
Sorted Z-values of atoh1a+atoh1b+atoh1c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of atoh1a+atoh1b+atoh1c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_27346176 | 1.15 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr15_-_23711689 | 1.03 |
ENSDART00000128644
|
ckmb
|
creatine kinase, muscle b |
| chr9_-_7695437 | 0.83 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr6_-_9964616 | 0.71 |
|
|
|
| chr17_+_27383737 | 0.69 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr18_-_6494014 | 0.66 |
ENSDART00000062423
|
tnni1c
|
troponin I, skeletal, slow c |
| chr9_-_23081250 | 0.66 |
|
|
|
| chr9_-_49834232 | 0.60 |
|
|
|
| chr25_+_7391458 | 0.59 |
ENSDART00000042928
|
fuk
|
fucokinase |
| chr10_-_43826919 | 0.59 |
ENSDART00000039551
ENSDART00000160786 ENSDART00000099134 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr23_-_18131105 | 0.59 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr24_+_7607915 | 0.58 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr8_-_6899611 | 0.58 |
ENSDART00000139545
|
wdr13
|
WD repeat domain 13 |
| chr8_-_22265875 | 0.57 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr20_+_34248780 | 0.56 |
ENSDART00000152870
|
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr23_-_18131209 | 0.55 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr22_+_34738389 | 0.55 |
ENSDART00000154372
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr10_-_38206238 | 0.55 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr2_+_2895642 | 0.54 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr18_+_30391910 | 0.53 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr8_+_7340538 | 0.53 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr1_+_51189343 | 0.52 |
ENSDART00000150757
|
BX548032.3
|
ENSDARG00000096092 |
| chr5_-_47656197 | 0.50 |
|
|
|
| chr5_+_29327479 | 0.50 |
|
|
|
| chr9_+_14052160 | 0.49 |
ENSDART00000124267
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr21_-_23710216 | 0.49 |
ENSDART00000132570
|
ncam1a
|
neural cell adhesion molecule 1a |
| chr8_+_48106836 | 0.49 |
ENSDART00000138717
|
si:ch211-263k4.2
|
si:ch211-263k4.2 |
| chr18_+_36825511 | 0.49 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr10_-_23939618 | 0.49 |
|
|
|
| chr8_-_14113377 | 0.49 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr18_+_23198330 | 0.48 |
ENSDART00000143977
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr24_-_23639458 | 0.48 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr6_+_46307828 | 0.48 |
ENSDART00000154817
|
si:dkeyp-67f1.1
|
si:dkeyp-67f1.1 |
| chr11_+_6893104 | 0.47 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr5_-_41672394 | 0.47 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr13_-_36265504 | 0.47 |
ENSDART00000140243
|
actn1
|
actinin, alpha 1 |
| chr3_+_32657833 | 0.46 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr24_+_24924379 | 0.46 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr19_-_34328673 | 0.45 |
ENSDART00000164563
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| KN149932v1_+_27584 | 0.45 |
|
|
|
| chr12_+_16845837 | 0.45 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr3_+_40667131 | 0.45 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr1_-_28241500 | 0.45 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr19_+_1743359 | 0.45 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr1_-_15814788 | 0.45 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr23_+_29981525 | 0.44 |
ENSDART00000035755
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr9_-_44493074 | 0.44 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr3_-_23466232 | 0.43 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr5_-_51172369 | 0.43 |
ENSDART00000164267
ENSDART00000157866 |
homer1b
|
homer scaffolding protein 1b |
| chr25_-_20160885 | 0.43 |
ENSDART00000160700
|
dnm1l
|
dynamin 1-like |
| chr2_-_44402486 | 0.42 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr11_-_25803101 | 0.42 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr23_+_35964754 | 0.42 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr15_+_9351511 | 0.42 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr21_+_9483423 | 0.41 |
ENSDART00000162834
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr11_-_32461160 | 0.41 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr2_-_42279180 | 0.41 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr10_+_34377959 | 0.41 |
|
|
|
| chr2_-_21512403 | 0.40 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr10_-_9123386 | 0.40 |
ENSDART00000149493
|
mocs2
|
molybdenum cofactor synthesis 2 |
| chr5_+_29327363 | 0.40 |
|
|
|
| chr20_+_6783810 | 0.40 |
ENSDART00000169966
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr5_+_37254520 | 0.40 |
ENSDART00000051222
|
ins
|
preproinsulin |
| chr10_+_15450466 | 0.40 |
ENSDART00000003839
|
nln
|
neurolysin (metallopeptidase M3 family) |
| chr16_+_3090170 | 0.39 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr10_+_15819127 | 0.39 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr21_-_19789339 | 0.38 |
|
|
|
| chr2_+_32033176 | 0.38 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr21_+_5868923 | 0.38 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr24_-_32750010 | 0.38 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr10_-_38206060 | 0.38 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr9_+_25285612 | 0.38 |
ENSDART00000135074
|
lrch1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr7_+_24617687 | 0.38 |
ENSDART00000173994
|
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr15_-_8541318 | 0.38 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr14_-_36523075 | 0.38 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr9_+_23854967 | 0.37 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr3_-_27944852 | 0.37 |
ENSDART00000122037
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_+_7014241 | 0.37 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr24_-_18540597 | 0.37 |
ENSDART00000176165
|
zgc:66014
|
zgc:66014 |
| chr6_-_9329917 | 0.37 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr1_-_24297477 | 0.36 |
|
|
|
| chr6_-_35049344 | 0.36 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr16_+_5625301 | 0.36 |
|
|
|
| chr1_+_21244242 | 0.36 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr8_-_17130805 | 0.35 |
|
|
|
| chr10_-_17383380 | 0.35 |
|
|
|
| chr10_-_37098396 | 0.35 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr8_-_18547147 | 0.35 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr11_+_41802482 | 0.35 |
ENSDART00000173022
|
camta1
|
calmodulin binding transcription activator 1 |
| chr10_+_15819085 | 0.35 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr7_-_71929355 | 0.35 |
|
|
|
| chr10_-_38206132 | 0.35 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr25_+_29873412 | 0.35 |
ENSDART00000067056
|
ticrr
|
TopBP1-interacting, checkpoint, and replication regulator |
| chr16_-_10404109 | 0.35 |
|
|
|
| chr22_+_16282153 | 0.35 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr5_+_29327447 | 0.34 |
|
|
|
| chr13_+_28723589 | 0.34 |
|
|
|
| chr18_+_8943793 | 0.34 |
ENSDART00000144247
|
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr2_-_44402452 | 0.34 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr12_+_5154886 | 0.34 |
ENSDART00000163897
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr2_+_32033028 | 0.33 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr18_-_3244618 | 0.33 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr8_+_29733109 | 0.33 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr10_-_17383466 | 0.33 |
|
|
|
| chr3_-_15889742 | 0.33 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr23_+_43284714 | 0.33 |
|
|
|
| chr6_-_6835861 | 0.33 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr7_-_55406347 | 0.32 |
ENSDART00000021009
|
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr5_-_21520111 | 0.32 |
|
|
|
| chr3_-_32471212 | 0.32 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr22_+_26543146 | 0.32 |
|
|
|
| chr14_-_23440160 | 0.31 |
ENSDART00000005842
|
fgf1a
|
fibroblast growth factor 1a |
| chr17_+_8874210 | 0.31 |
|
|
|
| chr16_+_5625552 | 0.31 |
|
|
|
| chr17_-_15659244 | 0.31 |
ENSDART00000142972
|
manea
|
mannosidase, endo-alpha |
| chr5_+_37144384 | 0.31 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr22_-_10367866 | 0.31 |
ENSDART00000142886
|
nisch
|
nischarin |
| chr7_+_40972616 | 0.30 |
ENSDART00000173814
|
scrib
|
scribbled planar cell polarity protein |
| chr4_+_17550101 | 0.30 |
ENSDART00000013313
|
itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr20_-_39371023 | 0.30 |
ENSDART00000127173
|
clu
|
clusterin |
| chr17_-_7990960 | 0.30 |
|
|
|
| chr18_-_47026460 | 0.30 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr18_+_29166912 | 0.30 |
ENSDART00000089031
|
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr18_-_12889296 | 0.30 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr10_-_13220838 | 0.30 |
ENSDART00000160265
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr9_+_33177527 | 0.30 |
ENSDART00000174533
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr14_+_2969679 | 0.30 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr7_-_19116999 | 0.29 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr5_+_43365449 | 0.29 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr2_-_17443642 | 0.29 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr25_-_18850809 | 0.29 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr7_-_68629353 | 0.29 |
|
|
|
| chr3_-_31713951 | 0.29 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr20_+_2443682 | 0.29 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr10_-_28474085 | 0.29 |
|
|
|
| chr1_+_58029993 | 0.28 |
|
|
|
| chr13_-_36265476 | 0.28 |
ENSDART00000133740
ENSDART00000100217 |
actn1
|
actinin, alpha 1 |
| chr15_-_7622212 | 0.28 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr5_-_33169062 | 0.28 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr4_-_4825948 | 0.27 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr24_+_7608106 | 0.27 |
ENSDART00000082467
|
cavin1b
|
caveolae associated protein 1b |
| chr7_+_72128846 | 0.27 |
ENSDART00000123887
|
MAPK8IP1 (1 of many)
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr25_-_28932291 | 0.27 |
ENSDART00000073488
|
CU469568.1
|
ENSDARG00000051805 |
| chr1_-_44942047 | 0.27 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr14_-_24304373 | 0.27 |
|
|
|
| chr8_+_52633069 | 0.26 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr19_-_41482388 | 0.26 |
ENSDART00000111982
|
sgce
|
sarcoglycan, epsilon |
| chr20_-_26102193 | 0.26 |
ENSDART00000136518
|
capn3b
|
calpain 3b |
| chr6_+_23835205 | 0.26 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr21_-_21336603 | 0.25 |
ENSDART00000079629
|
ppm1nb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Nb (putative) |
| chr24_+_7607852 | 0.25 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr13_+_25298383 | 0.25 |
|
|
|
| chr17_-_37040694 | 0.25 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr5_+_3965645 | 0.25 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr13_+_51154848 | 0.25 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr17_+_26593564 | 0.25 |
ENSDART00000159811
|
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr23_+_34103987 | 0.25 |
ENSDART00000175235
|
sars
|
seryl-tRNA synthetase |
| chr7_+_28862183 | 0.25 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr20_-_26102273 | 0.24 |
ENSDART00000063177
|
capn3b
|
calpain 3b |
| chr1_+_9999665 | 0.24 |
ENSDART00000054879
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr23_-_1008307 | 0.24 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr16_+_21122024 | 0.24 |
|
|
|
| chr7_+_6517248 | 0.24 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr12_-_14964315 | 0.24 |
ENSDART00000141909
|
pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr14_-_136402 | 0.24 |
|
|
|
| chr10_-_8764361 | 0.24 |
|
|
|
| chr6_-_12060971 | 0.24 |
ENSDART00000155685
|
pkp4
|
plakophilin 4 |
| chr20_+_51918693 | 0.24 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr23_+_39803084 | 0.24 |
ENSDART00000113893
|
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr1_-_40208469 | 0.24 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr2_-_21512154 | 0.24 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr22_+_16279764 | 0.23 |
|
|
|
| chr2_+_13779498 | 0.23 |
ENSDART00000149309
|
cfap57
|
cilia and flagella associated protein 57 |
| chr5_+_27560610 | 0.23 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr10_-_34927807 | 0.23 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_27667972 | 0.23 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr16_-_33976031 | 0.23 |
ENSDART00000110743
|
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr16_-_22397159 | 0.23 |
|
|
|
| chr10_+_21796176 | 0.22 |
ENSDART00000128215
|
CR774179.1
|
ENSDARG00000086969 |
| chr23_-_8797997 | 0.22 |
ENSDART00000014222
|
tcea2
|
transcription elongation factor A (SII), 2 |
| chr2_+_30266427 | 0.22 |
ENSDART00000135171
|
tmem70
|
transmembrane protein 70 |
| chr13_+_24148390 | 0.22 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr18_-_12889369 | 0.22 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr2_+_39142628 | 0.22 |
ENSDART00000179419
|
clstn2
|
calsyntenin 2 |
| chr16_+_46327528 | 0.22 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr24_+_38267919 | 0.22 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr17_+_44896438 | 0.22 |
ENSDART00000133103
|
coq6
|
coenzyme Q6 monooxygenase |
| chr19_+_24334831 | 0.22 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr8_+_26210913 | 0.22 |
ENSDART00000155529
|
wdr6
|
WD repeat domain 6 |
| chr5_+_56649893 | 0.21 |
ENSDART00000036331
|
atp6v1g1
|
ATPase, H+ transporting, lysosomal, V1 subunit G1 |
| chr18_+_25016753 | 0.21 |
ENSDART00000099476
|
fam174b
|
family with sequence similarity 174, member B |
| chr1_+_2074153 | 0.21 |
ENSDART00000058877
|
rap2ab
|
RAP2A, member of RAS oncogene family b |
| chr21_-_32449627 | 0.21 |
|
|
|
| chr10_+_29425917 | 0.21 |
|
|
|
| chr5_+_3172101 | 0.21 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr5_-_40133824 | 0.21 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr7_-_26653672 | 0.21 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr5_-_52357382 | 0.21 |
ENSDART00000157978
|
mrps30
|
mitochondrial ribosomal protein S30 |
| chr3_+_48362748 | 0.21 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr9_+_14052296 | 0.20 |
ENSDART00000124267
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 0.8 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.2 | 0.6 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.2 | 1.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.4 | GO:0001504 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.1 | 0.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.4 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.1 | 1.0 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.4 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.1 | 2.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0072171 | ureteric bud development(GO:0001657) branching involved in ureteric bud morphogenesis(GO:0001658) mesonephros development(GO:0001823) positive regulation of steroid metabolic process(GO:0045940) regulation of steroid biosynthetic process(GO:0050810) ureteric bud morphogenesis(GO:0060675) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) mesonephric tubule morphogenesis(GO:0072171) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.4 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.4 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.3 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.4 | GO:0043279 | response to alkaloid(GO:0043279) |
| 0.1 | 0.4 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.6 | GO:0045667 | skeletal muscle myosin thick filament assembly(GO:0030241) regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.3 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.1 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.1 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.4 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:1903895 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.6 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0010482 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:1901906 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.2 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.7 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.7 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 1.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.5 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 1.0 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.7 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.5 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0051429 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 3.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |