Project
DANIO-CODE
Navigation
Downloads
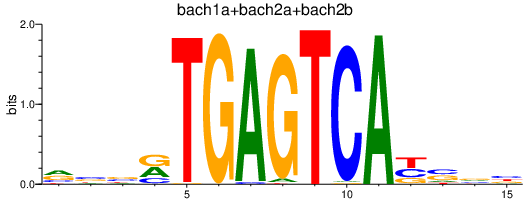
Results for bach1a+bach2a+bach2b
Z-value: 1.16
Transcription factors associated with bach1a+bach2a+bach2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bach2b
|
ENSDARG00000004074 | BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
|
bach2a
|
ENSDARG00000036569 | BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
|
bach1a
|
ENSDARG00000062553 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bach2b | dr10_dc_chr20_-_24223659_24223771 | -0.96 | 8.7e-09 | Click! |
| bach2a | dr10_dc_chr17_+_15666420_15666424 | 0.91 | 1.1e-06 | Click! |
| bach1a | dr10_dc_chr15_-_8215797_8215803 | 0.55 | 2.7e-02 | Click! |
Activity profile of bach1a+bach2a+bach2b motif
Sorted Z-values of bach1a+bach2a+bach2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of bach1a+bach2a+bach2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_45907269 | 5.55 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr23_+_2786407 | 4.58 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr6_+_51713264 | 4.19 |
ENSDART00000146281
|
fam65c
|
family with sequence similarity 65, member C |
| chr2_+_15432208 | 4.19 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr2_+_15432130 | 3.63 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr11_+_37371675 | 3.32 |
ENSDART00000140502
|
sh2d5
|
SH2 domain containing 5 |
| chr5_+_26888744 | 3.00 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr17_+_19606608 | 2.88 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr1_+_45907424 | 2.64 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr22_+_24194444 | 2.35 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr24_-_21224371 | 2.32 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr24_-_21224397 | 2.30 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr5_-_46028578 | 2.24 |
|
|
|
| chr5_+_26888419 | 2.19 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr14_+_30433715 | 2.17 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr9_+_28329384 | 2.16 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr5_+_35186037 | 2.08 |
ENSDART00000127383
ENSDART00000022043 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr19_-_3547196 | 2.05 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr16_-_24916921 | 1.94 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr15_+_25554119 | 1.88 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr14_+_30434569 | 1.86 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr11_-_14044356 | 1.83 |
ENSDART00000085158
|
tmem259
|
transmembrane protein 259 |
| chr2_-_49155324 | 1.81 |
|
|
|
| chr24_-_21828216 | 1.80 |
ENSDART00000030592
|
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| KN150286v1_-_2942 | 1.80 |
|
|
|
| chr23_-_37010705 | 1.79 |
ENSDART00000134461
|
zgc:193690
|
zgc:193690 |
| chr15_+_17407036 | 1.76 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr12_-_27121226 | 1.71 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr19_+_10742944 | 1.69 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr8_-_18637047 | 1.69 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr5_+_33949469 | 1.69 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr3_+_48810347 | 1.68 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr23_+_25929969 | 1.64 |
ENSDART00000145426
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr23_-_4769182 | 1.62 |
ENSDART00000144536
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr5_-_56810425 | 1.55 |
ENSDART00000147875
ENSDART00000142776 |
ENSDARG00000095415
|
ENSDARG00000095415 |
| chr16_+_46443800 | 1.55 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr8_+_48976657 | 1.53 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr1_+_46474561 | 1.46 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr12_-_36113358 | 1.46 |
|
|
|
| chr10_+_32107014 | 1.44 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr22_-_6532901 | 1.41 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr3_-_29430586 | 1.40 |
|
|
|
| chr23_-_4769010 | 1.39 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr11_-_22210506 | 1.39 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr7_+_38408691 | 1.36 |
ENSDART00000171691
|
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr19_-_44161905 | 1.34 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr7_+_26378093 | 1.34 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr15_-_43954665 | 1.32 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr22_+_31073990 | 1.32 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr13_+_40347099 | 1.30 |
|
|
|
| chr5_+_26888865 | 1.30 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr2_+_48090771 | 1.30 |
ENSDART00000135584
|
ftr19
|
finTRIM family, member 19 |
| chr3_-_3665840 | 1.29 |
ENSDART00000078886
ENSDART00000170539 |
ENSDARG00000008835
|
ENSDARG00000008835 |
| chr6_-_19473903 | 1.27 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr19_-_44161951 | 1.26 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr13_+_6908853 | 1.24 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr7_-_57055583 | 1.22 |
|
|
|
| chr11_-_38856849 | 1.21 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr5_+_44204845 | 1.21 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr19_+_43277870 | 1.20 |
ENSDART00000135164
|
pdcd6ip
|
programmed cell death 6 interacting protein |
| chr24_-_33475705 | 1.20 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr2_-_8002471 | 1.19 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr8_-_18637019 | 1.19 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr20_-_16949608 | 1.18 |
ENSDART00000027582
|
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr23_+_45486958 | 1.17 |
ENSDART00000111126
|
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr15_+_40340914 | 1.16 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr21_-_30217360 | 1.16 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr12_-_27121343 | 1.13 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr23_+_4315077 | 1.05 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr9_-_2522639 | 1.01 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr21_+_13136338 | 0.99 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr20_-_43826667 | 0.99 |
ENSDART00000100637
|
mixl1
|
Mix paired-like homeobox |
| chr13_-_2296283 | 0.99 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr6_-_6818607 | 0.98 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr1_-_51863300 | 0.98 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_-_30561224 | 0.98 |
ENSDART00000155436
|
CR848742.1
|
ENSDARG00000097142 |
| chr5_+_44204661 | 0.97 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr18_+_15873032 | 0.96 |
ENSDART00000141800
|
eea1
|
early endosome antigen 1 |
| chr2_+_37855019 | 0.96 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr13_-_37523664 | 0.95 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr13_-_2296041 | 0.95 |
ENSDART00000126697
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr23_+_17596460 | 0.95 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr18_+_44856321 | 0.95 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr20_-_16949680 | 0.93 |
ENSDART00000027582
|
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr7_+_71262845 | 0.92 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr14_+_28158771 | 0.91 |
ENSDART00000019003
|
psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr14_+_8641470 | 0.90 |
ENSDART00000158025
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr24_+_36351542 | 0.88 |
ENSDART00000062745
|
riok3
|
RIO kinase 3 (yeast) |
| chr14_+_8641516 | 0.86 |
ENSDART00000158025
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr3_+_17728616 | 0.85 |
ENSDART00000167953
|
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr13_+_7243353 | 0.85 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr19_+_37533306 | 0.85 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr12_-_3042545 | 0.85 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_+_7584201 | 0.82 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr19_-_42847112 | 0.81 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr7_+_34277682 | 0.80 |
ENSDART00000073447
|
fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_1102450 | 0.79 |
ENSDART00000115044
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr6_+_41455676 | 0.79 |
ENSDART00000007798
ENSDART00000162135 |
twf2a
|
twinfilin actin-binding protein 2a |
| chr7_-_29956782 | 0.78 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr5_-_62963536 | 0.78 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr4_-_4526152 | 0.77 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr8_+_48976994 | 0.77 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr4_+_76548332 | 0.77 |
ENSDART00000159452
ENSDART00000092415 |
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_-_41523801 | 0.75 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr5_-_41523490 | 0.75 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr7_+_7463565 | 0.74 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr5_-_32698092 | 0.73 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr23_-_37010664 | 0.73 |
ENSDART00000102886
|
zgc:193690
|
zgc:193690 |
| chr4_-_7644310 | 0.72 |
ENSDART00000157751
|
BX649411.1
|
ENSDARG00000101254 |
| chr2_-_1028487 | 0.71 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr8_-_49217824 | 0.70 |
|
|
|
| chr15_-_31296457 | 0.69 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr11_-_18649022 | 0.69 |
|
|
|
| chr15_-_31296524 | 0.67 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr12_-_3042573 | 0.65 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr19_-_37533194 | 0.64 |
|
|
|
| chr11_-_29689360 | 0.64 |
ENSDART00000145251
|
pir
|
pirin |
| chr16_+_27608996 | 0.64 |
ENSDART00000147611
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr9_+_22846535 | 0.64 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr20_-_26516874 | 0.63 |
|
|
|
| chr16_+_48772036 | 0.61 |
ENSDART00000148709
|
brd2b
|
bromodomain containing 2b |
| chr20_+_49380442 | 0.61 |
|
|
|
| chr15_+_20961370 | 0.61 |
ENSDART00000154854
|
CR848784.5
|
ENSDARG00000097996 |
| chr5_+_32698516 | 0.60 |
ENSDART00000097945
|
usp20
|
ubiquitin specific peptidase 20 |
| chr5_+_44205128 | 0.60 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr20_-_16649213 | 0.60 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr19_+_10477101 | 0.59 |
ENSDART00000151735
|
necap1
|
NECAP endocytosis associated 1 |
| chr13_+_7243325 | 0.59 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr9_-_30560440 | 0.58 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr17_+_28657998 | 0.56 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr6_-_39313358 | 0.55 |
|
|
|
| chr22_+_39012456 | 0.55 |
ENSDART00000176985
|
CABZ01044615.1
|
ENSDARG00000108810 |
| chr11_-_29689291 | 0.54 |
ENSDART00000145251
|
pir
|
pirin |
| chr8_+_48976926 | 0.50 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr7_-_29262814 | 0.47 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr3_-_34687314 | 0.47 |
ENSDART00000084448
|
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr11_+_19208363 | 0.46 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr21_-_30217568 | 0.45 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr8_-_46904610 | 0.44 |
|
|
|
| chr13_-_2296317 | 0.44 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr7_-_6273677 | 0.44 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr6_+_55250892 | 0.44 |
|
|
|
| chr21_-_20983956 | 0.43 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr19_-_8829701 | 0.43 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr20_-_34167039 | 0.42 |
ENSDART00000017941
|
tprb
|
translocated promoter region b, nuclear basket protein |
| chr13_+_21757676 | 0.42 |
ENSDART00000162709
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr6_+_37774863 | 0.42 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr17_-_20675997 | 0.42 |
ENSDART00000063613
ENSDART00000171575 |
ccdc6a
|
coiled-coil domain containing 6a |
| chr5_+_41463969 | 0.42 |
ENSDART00000035235
|
UBB
|
si:ch211-202a12.4 |
| chr21_-_10962154 | 0.41 |
ENSDART00000084061
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr4_-_1952230 | 0.41 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr1_+_46474845 | 0.41 |
ENSDART00000053152
|
cbr1
|
carbonyl reductase 1 |
| chr16_-_13927947 | 0.40 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr16_-_13927684 | 0.40 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr16_+_17706214 | 0.39 |
|
|
|
| chr22_+_39098205 | 0.38 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr3_+_12438915 | 0.38 |
ENSDART00000159252
|
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr10_+_187740 | 0.36 |
ENSDART00000167367
|
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr22_-_6532694 | 0.32 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr14_-_36422904 | 0.32 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr14_-_5509554 | 0.31 |
|
|
|
| chr4_-_7644374 | 0.31 |
ENSDART00000157751
|
BX649411.1
|
ENSDARG00000101254 |
| chr21_+_10773736 | 0.31 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr7_-_29452646 | 0.31 |
ENSDART00000112651
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr15_+_17407258 | 0.30 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr11_+_44805115 | 0.28 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| KN150359v1_+_69830 | 0.28 |
|
|
|
| chr17_+_41313762 | 0.26 |
ENSDART00000128482
|
fosl2
|
fos-like antigen 2 |
| chr16_-_27609082 | 0.26 |
|
|
|
| chr15_-_43954596 | 0.25 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr14_-_470112 | 0.25 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr10_-_28474052 | 0.25 |
|
|
|
| chr21_+_6009262 | 0.25 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr11_+_11284030 | 0.23 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr19_+_33506239 | 0.23 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr18_+_15872911 | 0.20 |
ENSDART00000141800
|
eea1
|
early endosome antigen 1 |
| chr12_+_8040369 | 0.20 |
ENSDART00000124084
|
zgc:194508
|
zgc:194508 |
| chr19_-_10476653 | 0.19 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr10_-_7019880 | 0.19 |
ENSDART00000123312
|
sh2d4a
|
SH2 domain containing 4A |
| chr18_-_15187432 | 0.18 |
ENSDART00000111333
|
mterf2
|
mitochondrial transcription termination factor 2 |
| chr7_+_71262935 | 0.18 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr3_+_12438761 | 0.17 |
ENSDART00000159252
|
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr17_+_51593393 | 0.17 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr22_+_17278690 | 0.16 |
ENSDART00000088460
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_-_30654776 | 0.16 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr11_+_6450876 | 0.16 |
ENSDART00000170180
|
CU062501.2
|
ENSDARG00000103967 |
| chr17_+_35149038 | 0.16 |
ENSDART00000163557
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr4_+_46414646 | 0.16 |
ENSDART00000179615
|
BX324155.2
|
ENSDARG00000107359 |
| chr5_-_42350607 | 0.16 |
ENSDART00000149868
|
grsf1
|
G-rich RNA sequence binding factor 1 |
| chr20_-_43826581 | 0.14 |
ENSDART00000100637
|
mixl1
|
Mix paired-like homeobox |
| chr17_-_31027949 | 0.14 |
ENSDART00000152016
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr5_-_2540097 | 0.14 |
|
|
|
| chr1_+_51503405 | 0.13 |
ENSDART00000171597
ENSDART00000007079 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr11_-_269181 | 0.13 |
|
|
|
| chr13_-_49294664 | 0.13 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr21_+_22856217 | 0.12 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr2_-_37854616 | 0.12 |
ENSDART00000171409
|
mettl17
|
methyltransferase like 17 |
| chr23_+_24575330 | 0.12 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr22_+_39110524 | 0.11 |
ENSDART00000153841
|
lmcd1
|
LIM and cysteine-rich domains 1 |
| KN150266v1_-_69477 | 0.10 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.5 | 2.8 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.4 | 2.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.4 | 4.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.3 | 0.9 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 1.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 0.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 1.8 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.2 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 3.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 6.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 1.1 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.2 | 2.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.2 | 2.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.5 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 7.8 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.2 | 1.8 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 1.5 | GO:0035283 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 2.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.5 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 2.8 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 0.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.8 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.1 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 3.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.7 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.3 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 1.6 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.6 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 3.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.2 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 2.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:1901906 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.4 | GO:0030202 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.0 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 2.8 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.4 | GO:0055088 | lipid homeostasis(GO:0055088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 2.4 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.4 | 1.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 1.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 1.7 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 2.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 4.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 3.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0030665 | clathrin vesicle coat(GO:0030125) clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 4.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 3.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.6 | 2.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 2.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 8.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 1.7 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.3 | 0.8 | GO:0016844 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.2 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 2.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 2.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 2.9 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 6.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 6.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.4 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.1 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 2.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 3.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.2 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.2 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |