Project
DANIO-CODE
Navigation
Downloads
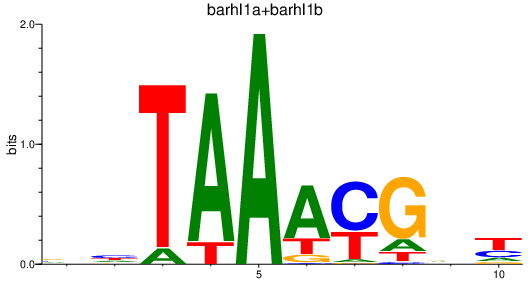
Results for barhl1a+barhl1b
Z-value: 2.61
Transcription factors associated with barhl1a+barhl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl1b
|
ENSDARG00000019013 | BarH-like homeobox 1b |
|
barhl1a
|
ENSDARG00000035508 | BarH-like homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl1a | dr10_dc_chr5_-_29150278_29150288 | -0.90 | 2.4e-06 | Click! |
Activity profile of barhl1a+barhl1b motif
Sorted Z-values of barhl1a+barhl1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl1a+barhl1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_44628278 | 9.36 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr6_+_23476669 | 7.69 |
|
|
|
| chr2_-_32404106 | 6.57 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr14_+_32578103 | 5.40 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr2_-_2373601 | 5.38 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr7_+_24855467 | 5.31 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr14_+_32578253 | 5.13 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr12_+_23543100 | 4.89 |
ENSDART00000111334
|
mtpap
|
mitochondrial poly(A) polymerase |
| chr23_-_37553481 | 4.85 |
|
|
|
| chr16_+_47283253 | 4.84 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr23_-_35691369 | 4.62 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr5_-_56953587 | 4.58 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr10_+_6925373 | 4.47 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr5_+_24016742 | 4.43 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr14_+_22201617 | 4.04 |
ENSDART00000174265
|
smyd5
|
SMYD family member 5 |
| chr5_-_23211957 | 4.01 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr25_-_3619590 | 3.91 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr19_-_10411573 | 3.91 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr25_-_7794186 | 3.84 |
ENSDART00000104686
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr5_-_56953716 | 3.75 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr23_+_17583666 | 3.62 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr9_+_41658126 | 3.59 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr2_-_23734866 | 3.51 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr16_+_47283374 | 3.46 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr6_+_48349630 | 3.43 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr23_+_40517744 | 3.42 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr6_+_277714 | 3.41 |
|
|
|
| chr16_-_34240818 | 3.41 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr23_+_39961676 | 3.39 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| chr22_-_31089831 | 3.39 |
|
|
|
| chr13_-_35782121 | 3.39 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr1_-_44628181 | 3.38 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr9_+_8990576 | 3.27 |
ENSDART00000133899
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr7_+_15479700 | 3.24 |
|
|
|
| chr17_+_7377230 | 3.23 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr20_-_31035185 | 3.22 |
ENSDART00000147359
|
wtap
|
WT1 associated protein |
| chr17_+_22291546 | 3.19 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr7_+_36196295 | 3.13 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr23_-_6831711 | 3.11 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr18_+_6396038 | 3.05 |
ENSDART00000024576
|
fkbp4
|
FK506 binding protein 4 |
| chr16_+_11888601 | 3.04 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr6_+_277946 | 3.04 |
|
|
|
| chr2_-_29939721 | 3.02 |
ENSDART00000031130
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr7_+_23809924 | 3.01 |
ENSDART00000141749
|
acin1b
|
apoptotic chromatin condensation inducer 1b |
| chr20_+_35535503 | 2.98 |
ENSDART00000153249
|
tdrd6
|
tudor domain containing 6 |
| chr22_+_3994627 | 2.96 |
ENSDART00000166768
|
timm44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr23_-_17055330 | 2.91 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr25_-_36512943 | 2.89 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr5_-_27549500 | 2.89 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr23_+_17583383 | 2.81 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr2_+_56534374 | 2.78 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr7_-_18460264 | 2.75 |
ENSDART00000173905
|
trmt10a
|
tRNA methyltransferase 10A |
| chr17_-_31802796 | 2.75 |
ENSDART00000172519
|
abraxas2b
|
abraxas 2b, BRISC complex subunit |
| chr6_+_3313468 | 2.74 |
ENSDART00000011785
|
rad54l
|
RAD54-like (S. cerevisiae) |
| chr5_-_19342462 | 2.73 |
ENSDART00000159309
|
trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr17_+_28653205 | 2.68 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr21_+_25764848 | 2.67 |
ENSDART00000136146
ENSDART00000035062 |
nf2b
|
neurofibromin 2b (merlin) |
| chr18_+_6396115 | 2.66 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr23_-_41367783 | 2.66 |
|
|
|
| chr16_+_25370991 | 2.65 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr18_+_8959686 | 2.60 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr14_-_16469435 | 2.60 |
ENSDART00000113711
|
zgc:123335
|
zgc:123335 |
| chr8_-_16638825 | 2.55 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr23_-_25853364 | 2.55 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr1_-_8843231 | 2.53 |
ENSDART00000110790
|
si:ch73-12o23.1
|
si:ch73-12o23.1 |
| chr5_+_22006830 | 2.53 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr9_-_21677941 | 2.51 |
ENSDART00000121939
ENSDART00000080404 |
mphosph8
|
M-phase phosphoprotein 8 |
| chr7_+_28341426 | 2.49 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr2_+_27730940 | 2.46 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr21_+_26954898 | 2.46 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr13_+_46429054 | 2.45 |
ENSDART00000160401
|
tmem63ba
|
transmembrane protein 63Ba |
| chr2_-_23735018 | 2.45 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr19_-_6925090 | 2.45 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr3_-_36080647 | 2.43 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr6_-_19813235 | 2.41 |
ENSDART00000151152
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr2_-_32403565 | 2.39 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr22_-_10026579 | 2.39 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr21_-_25765319 | 2.38 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr25_-_7794262 | 2.35 |
ENSDART00000156761
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr19_+_42486563 | 2.34 |
ENSDART00000151605
ENSDART00000166422 |
nfyc
|
nuclear transcription factor Y, gamma |
| chr3_+_21905456 | 2.34 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr15_-_19192862 | 2.33 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr15_-_970988 | 2.33 |
ENSDART00000106627
ENSDART00000102239 |
zgc:162936
|
zgc:162936 |
| chr21_-_41602067 | 2.32 |
|
|
|
| chr25_+_19443128 | 2.30 |
ENSDART00000141056
|
nedd1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr3_+_40142679 | 2.27 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr4_+_13413303 | 2.26 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr7_-_33965017 | 2.26 |
|
|
|
| chr16_-_21064435 | 2.25 |
ENSDART00000040727
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| KN150604v1_-_7321 | 2.25 |
|
|
|
| chr7_+_37989122 | 2.24 |
ENSDART00000143611
|
cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr23_+_40240121 | 2.23 |
|
|
|
| chr7_+_21485734 | 2.22 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr2_-_32842618 | 2.22 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr7_+_24374240 | 2.22 |
ENSDART00000087691
|
gba2
|
glucosidase, beta (bile acid) 2 |
| chr8_+_25742461 | 2.20 |
ENSDART00000062403
|
tmem9
|
transmembrane protein 9 |
| chr16_-_42202425 | 2.19 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr13_+_7055611 | 2.18 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr9_+_41658313 | 2.18 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr22_-_52569 | 2.18 |
|
|
|
| chr20_-_32246045 | 2.17 |
ENSDART00000048537
ENSDART00000152984 |
cep57l1
|
centrosomal protein 57, like 1 |
| chr22_-_10744481 | 2.16 |
|
|
|
| chr17_-_17739301 | 2.15 |
ENSDART00000157128
|
adck1
|
aarF domain containing kinase 1 |
| chr17_+_24427297 | 2.15 |
ENSDART00000163221
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr14_-_6096258 | 2.14 |
|
|
|
| chr13_-_23888383 | 2.14 |
ENSDART00000026189
|
cog2
|
component of oligomeric golgi complex 2 |
| chr5_+_23582676 | 2.14 |
ENSDART00000139520
|
tp53
|
tumor protein p53 |
| chr5_-_32290516 | 2.13 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr4_+_15007453 | 2.12 |
ENSDART00000146156
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr2_+_32842978 | 2.11 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr18_+_22297627 | 2.11 |
ENSDART00000151919
|
ctcf
|
CCCTC-binding factor (zinc finger protein) |
| chr3_+_30187036 | 2.11 |
ENSDART00000151006
|
CR936968.1
|
ENSDARG00000096295 |
| chr7_-_52850565 | 2.08 |
|
|
|
| chr10_+_2554651 | 2.06 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin-like 2 |
| chr4_-_9349572 | 2.05 |
ENSDART00000148155
|
si:ch211-125a15.1
|
si:ch211-125a15.1 |
| chr17_+_28653236 | 2.04 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr14_+_21522552 | 2.03 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr7_-_40359613 | 2.03 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr13_+_10940798 | 2.03 |
ENSDART00000114911
|
sdccag8
|
serologically defined colon cancer antigen 8 |
| chr23_+_37476457 | 2.02 |
ENSDART00000178064
|
AL954146.3
|
ENSDARG00000108629 |
| chr16_+_26858575 | 2.00 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr1_+_43472177 | 2.00 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr3_+_42667814 | 1.99 |
ENSDART00000172425
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr23_+_31669909 | 1.98 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr25_-_3221372 | 1.98 |
ENSDART00000169418
|
golt1bb
|
golgi transport 1Bb |
| chr21_+_37405864 | 1.98 |
ENSDART00000148956
|
znf346
|
zinc finger protein 346 |
| chr6_-_6818607 | 1.97 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr16_+_11888545 | 1.97 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr15_-_30976483 | 1.96 |
ENSDART00000112511
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr21_+_21871674 | 1.94 |
|
|
|
| chr3_-_36940649 | 1.93 |
ENSDART00000110716
ENSDART00000160233 |
tubg1
|
tubulin, gamma 1 |
| chr16_+_25370955 | 1.92 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr19_-_10411518 | 1.91 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr18_-_20159345 | 1.91 |
|
|
|
| chr7_-_20212157 | 1.90 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr16_+_20250148 | 1.89 |
ENSDART00000146436
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr4_-_3014778 | 1.89 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr16_+_8067699 | 1.89 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr22_-_9866462 | 1.88 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr16_+_9509747 | 1.88 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr6_+_8417443 | 1.87 |
|
|
|
| chr22_-_26971170 | 1.85 |
ENSDART00000162742
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr11_-_40255430 | 1.85 |
ENSDART00000127932
|
miip
|
migration and invasion inhibitory protein |
| chr19_+_30210491 | 1.84 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr7_+_33965084 | 1.84 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr24_+_39339391 | 1.83 |
ENSDART00000168705
|
si:ch73-103b11.2
|
si:ch73-103b11.2 |
| chr20_-_44020837 | 1.83 |
ENSDART00000149897
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr10_-_35313462 | 1.81 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr18_+_8362265 | 1.80 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr10_-_42279805 | 1.80 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr21_-_36710989 | 1.78 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr15_-_14162320 | 1.78 |
ENSDART00000156947
|
si:ch211-116o3.5
|
si:ch211-116o3.5 |
| chr3_-_52428451 | 1.77 |
ENSDART00000155248
|
si:dkey-210j14.5
|
si:dkey-210j14.5 |
| chr6_+_59580554 | 1.76 |
|
|
|
| chr20_-_31035087 | 1.76 |
ENSDART00000147359
|
wtap
|
WT1 associated protein |
| chr5_+_7691621 | 1.75 |
ENSDART00000161261
|
lmbrd2a
|
LMBR1 domain containing 2a |
| chr3_+_34876160 | 1.74 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr17_+_8596825 | 1.74 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr6_-_51771558 | 1.72 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr5_+_23583233 | 1.72 |
ENSDART00000177458
|
tp53
|
tumor protein p53 |
| chr5_-_32290272 | 1.72 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr17_-_33764220 | 1.71 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr4_-_9780251 | 1.69 |
ENSDART00000080744
|
svopl
|
SVOP-like |
| chr1_-_52833426 | 1.68 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr13_-_15668180 | 1.67 |
ENSDART00000124688
|
bag5
|
BCL2-associated athanogene 5 |
| chr21_+_43183835 | 1.67 |
ENSDART00000175107
|
aff4
|
AF4/FMR2 family, member 4 |
| chr13_-_23888408 | 1.67 |
ENSDART00000026189
|
cog2
|
component of oligomeric golgi complex 2 |
| chr3_-_29837017 | 1.67 |
ENSDART00000139310
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr22_+_1568354 | 1.66 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr17_-_20675997 | 1.66 |
ENSDART00000063613
ENSDART00000171575 |
ccdc6a
|
coiled-coil domain containing 6a |
| chr15_-_19192933 | 1.66 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr24_-_14447825 | 1.65 |
|
|
|
| chr1_-_30783409 | 1.65 |
ENSDART00000146891
|
cenpk
|
centromere protein K |
| chr23_+_17583294 | 1.65 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr6_-_54049582 | 1.64 |
|
|
|
| chr23_-_31986679 | 1.64 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr2_-_38097978 | 1.63 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr12_-_8920541 | 1.63 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr12_+_28795843 | 1.61 |
ENSDART00000076342
|
rnf40
|
ring finger protein 40 |
| chr2_-_38098516 | 1.60 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr16_+_29442108 | 1.59 |
ENSDART00000050535
|
rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr12_-_7412438 | 1.59 |
|
|
|
| chr18_+_22297501 | 1.58 |
|
|
|
| chr12_-_26939563 | 1.58 |
ENSDART00000076103
|
bc2
|
putative breast adenocarcinoma marker |
| chr1_+_28463888 | 1.57 |
ENSDART00000085648
|
lig4
|
ligase IV, DNA, ATP-dependent |
| chr6_+_28762398 | 1.57 |
ENSDART00000146574
|
tprg1
|
tumor protein p63 regulated 1 |
| chr3_+_54507075 | 1.57 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr5_-_48028931 | 1.57 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr11_+_25455772 | 1.55 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr21_+_18369660 | 1.55 |
|
|
|
| chr6_-_42339370 | 1.54 |
ENSDART00000075601
ENSDART00000125089 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr2_-_16780900 | 1.54 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr19_+_44073792 | 1.53 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr2_-_41035670 | 1.52 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr7_+_5789700 | 1.51 |
ENSDART00000173025
|
si:dkey-23a13.9
|
si:dkey-23a13.9 |
| chr21_+_25764787 | 1.50 |
ENSDART00000136146
ENSDART00000035062 |
nf2b
|
neurofibromin 2b (merlin) |
| chr20_-_32502517 | 1.50 |
ENSDART00000153411
|
afg1lb
|
AFG1 like ATPase b |
| chr19_+_30835769 | 1.50 |
ENSDART00000023074
|
mrps18b
|
mitochondrial ribosomal protein S18B |
| chr7_+_5789797 | 1.49 |
ENSDART00000173025
|
si:dkey-23a13.9
|
si:dkey-23a13.9 |
| chr7_-_49364447 | 1.48 |
ENSDART00000011456
|
tsg101b
|
tumor susceptibility 101b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 1.2 | 3.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 1.1 | 4.5 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.9 | 2.7 | GO:0097702 | response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.9 | 3.6 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.9 | 5.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.8 | 4.8 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.8 | 2.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.8 | 5.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.7 | 2.7 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.6 | 2.5 | GO:0032196 | transposition(GO:0032196) |
| 0.6 | 8.3 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.6 | 1.7 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.6 | 1.7 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.5 | 1.6 | GO:0090242 | mesodermal-endodermal cell signaling(GO:0003131) response to vitamin A(GO:0033189) cellular response to vitamin A(GO:0071299) retinoic acid receptor signaling pathway involved in somitogenesis(GO:0090242) |
| 0.5 | 2.1 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.5 | 4.5 | GO:0061458 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.5 | 1.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.5 | 1.8 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.5 | 2.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.5 | 2.3 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.5 | 9.6 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.4 | 4.4 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.4 | 15.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.4 | 4.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.4 | 1.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.4 | 3.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.4 | 1.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.4 | 2.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.4 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 3.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 3.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.3 | 4.0 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.3 | 1.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 2.2 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.3 | 1.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.3 | 8.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 2.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 2.5 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.3 | 3.0 | GO:0030719 | P granule organization(GO:0030719) |
| 0.3 | 3.4 | GO:1901673 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.3 | 2.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 2.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.4 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 1.0 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.2 | 5.1 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.2 | 3.8 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 1.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 2.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 4.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 4.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.2 | 1.7 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.2 | 0.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 1.8 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 4.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 0.6 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.4 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 0.4 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 3.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 0.6 | GO:0098719 | sodium ion import(GO:0097369) inorganic cation import into cell(GO:0098659) sodium ion import across plasma membrane(GO:0098719) inorganic ion import into cell(GO:0099587) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 3.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 2.6 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 3.3 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 2.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.4 | GO:0051754 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 2.1 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 4.5 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 3.3 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.1 | 2.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 1.6 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.1 | 0.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 4.4 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.1 | 2.5 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 6.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 2.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 1.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.2 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 1.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.5 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.2 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 2.5 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.8 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.9 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.3 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.3 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 2.4 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.3 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.5 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.4 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.4 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0008623 | CHRAC(GO:0008623) |
| 1.0 | 3.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.9 | 2.7 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.9 | 2.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.8 | 8.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.8 | 5.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.7 | 4.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.6 | 4.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.6 | 3.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 1.8 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.5 | 1.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.5 | 3.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 1.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 2.0 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 7.9 | GO:0043186 | P granule(GO:0043186) |
| 0.3 | 5.3 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.3 | 2.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 1.0 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.3 | 3.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 0.9 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.2 | 2.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 5.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 1.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.6 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 3.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 5.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 2.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 7.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.3 | GO:0005764 | lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.2 | 3.7 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 1.2 | 4.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 1.2 | 3.6 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 1.1 | 6.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.7 | 2.9 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.7 | 2.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.7 | 2.7 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.6 | 2.5 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.6 | 1.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.6 | 1.7 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 1.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.4 | 1.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 1.5 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.4 | 1.1 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.3 | 2.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 1.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 2.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.3 | 2.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 1.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 1.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 1.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 1.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 2.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 3.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 1.8 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.7 | GO:0035258 | histone kinase activity(GO:0035173) steroid hormone receptor binding(GO:0035258) |
| 0.2 | 1.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 2.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 16.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 4.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 1.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 2.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 4.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 2.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.4 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 2.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 7.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 3.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.4 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 2.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 2.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 19.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 2.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.4 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 2.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 4.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 3.1 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 3.8 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.0 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.4 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 3.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.3 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 6.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 7.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.5 | 4.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 4.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.4 | 3.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 2.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 1.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 2.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.3 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.0 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |