Project
DANIO-CODE
Navigation
Downloads
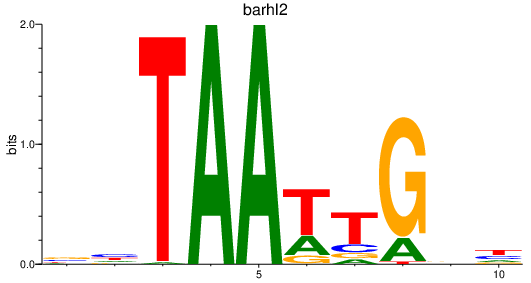
Results for barhl2
Z-value: 0.91
Transcription factors associated with barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl2
|
ENSDARG00000104361 | BarH-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl2 | dr10_dc_chr6_+_24717985_24718056 | -0.83 | 6.7e-05 | Click! |
Activity profile of barhl2 motif
Sorted Z-values of barhl2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25740782 | 3.32 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr22_-_20670164 | 2.97 |
ENSDART00000169077
|
org
|
oogenesis-related gene |
| chr18_-_40718244 | 2.74 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr17_-_30884905 | 2.50 |
ENSDART00000131633
ENSDART00000146824 |
evla
|
Enah/Vasp-like a |
| KN150266v1_-_68652 | 2.32 |
|
|
|
| chr6_+_23476669 | 2.26 |
|
|
|
| chr11_-_6442588 | 2.21 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr2_-_32404106 | 2.15 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr17_-_45021393 | 2.13 |
|
|
|
| chr17_+_16038358 | 1.97 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr8_+_45326435 | 1.97 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr16_-_24727689 | 1.96 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr19_-_20819477 | 1.94 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr10_+_6925373 | 1.75 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr3_+_42395984 | 1.75 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr17_+_16421892 | 1.73 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr23_-_35691369 | 1.71 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr1_-_18118467 | 1.65 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr11_-_44539778 | 1.58 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr19_+_21209328 | 1.56 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr12_-_28680035 | 1.56 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr24_-_9844547 | 1.56 |
|
|
|
| chr17_+_50622734 | 1.54 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr13_+_46652067 | 1.47 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr8_-_44247277 | 1.44 |
|
|
|
| chr20_-_23527234 | 1.43 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_16038103 | 1.41 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_+_29052293 | 1.41 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr1_-_54570813 | 1.40 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr10_-_44713495 | 1.38 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr11_+_25302180 | 1.38 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr8_+_41003546 | 1.37 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_+_6341404 | 1.35 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr5_+_29193876 | 1.34 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr1_+_9443056 | 1.31 |
ENSDART00000110698
|
rbm46
|
RNA binding motif protein 46 |
| chr21_-_2292663 | 1.29 |
ENSDART00000164015
|
zgc:66483
|
zgc:66483 |
| chr8_-_28323975 | 1.27 |
|
|
|
| chr5_-_56953587 | 1.22 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr21_-_3548863 | 1.20 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr11_-_6979695 | 1.16 |
ENSDART00000173242
ENSDART00000172896 ENSDART00000102493 |
zgc:173548
|
zgc:173548 |
| chr9_-_35824470 | 1.15 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr18_+_8959686 | 1.15 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| KN150266v1_-_68620 | 1.15 |
|
|
|
| chr5_-_56953716 | 1.13 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr22_-_746251 | 1.13 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr23_+_28396415 | 1.12 |
ENSDART00000142179
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr24_+_10758290 | 1.10 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr7_+_27184121 | 1.08 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr13_-_36785584 | 1.08 |
ENSDART00000167154
|
trim9
|
tripartite motif containing 9 |
| chr18_+_6417959 | 1.08 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr11_+_25302072 | 1.06 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr17_+_7377230 | 1.06 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr3_-_20989306 | 1.06 |
|
|
|
| chr10_-_34971926 | 1.05 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr20_+_38354928 | 1.05 |
ENSDART00000149160
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr23_+_40517744 | 1.03 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr20_-_3220106 | 1.02 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr3_-_40142513 | 1.00 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr11_+_25302044 | 1.00 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr6_-_12016481 | 0.99 |
|
|
|
| chr7_+_15479700 | 0.99 |
|
|
|
| chr3_+_40142679 | 0.99 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr16_+_42567707 | 0.99 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr2_-_26820425 | 0.97 |
ENSDART00000132125
|
acadm
|
acyl-CoA dehydrogenase, C-4 to C-12 straight chain |
| chr14_+_29932533 | 0.97 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr8_-_53165501 | 0.95 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr24_+_23571714 | 0.95 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr20_+_14893184 | 0.93 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr24_-_9874069 | 0.92 |
ENSDART00000176344
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr7_-_26332194 | 0.92 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr20_+_18803841 | 0.90 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr3_-_19890717 | 0.89 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr25_-_24105014 | 0.88 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr11_+_11319810 | 0.88 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr1_-_7882397 | 0.87 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr7_+_24257251 | 0.86 |
ENSDART00000136473
|
ENSDARG00000079281
|
ENSDARG00000079281 |
| chr12_-_10529153 | 0.86 |
ENSDART00000047769
|
myof
|
myoferlin |
| chr16_+_47283253 | 0.86 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr1_+_21244200 | 0.86 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr19_-_35286549 | 0.84 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr1_+_18118735 | 0.83 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr14_+_50010976 | 0.82 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr12_+_23543100 | 0.81 |
ENSDART00000111334
|
mtpap
|
mitochondrial poly(A) polymerase |
| chr15_-_6969345 | 0.81 |
ENSDART00000169529
|
mrps22
|
mitochondrial ribosomal protein S22 |
| chr15_-_25333773 | 0.81 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr18_-_43890836 | 0.80 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr3_-_26112995 | 0.78 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr7_+_35163845 | 0.78 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr17_+_1799173 | 0.78 |
ENSDART00000112183
|
cep170b
|
centrosomal protein 170B |
| chr10_-_32550351 | 0.76 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr4_+_13902603 | 0.76 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr5_-_53907908 | 0.76 |
ENSDART00000158069
|
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr23_-_6831711 | 0.76 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr20_+_46668743 | 0.75 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr10_-_36848872 | 0.75 |
|
|
|
| chr17_-_12467563 | 0.75 |
|
|
|
| chr10_-_44713414 | 0.75 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr16_-_23975775 | 0.74 |
ENSDART00000077807
|
BX004983.1
|
ENSDARG00000055446 |
| chr16_-_13790348 | 0.74 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr7_+_28453707 | 0.74 |
ENSDART00000173947
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr3_+_42396136 | 0.73 |
ENSDART00000165708
|
micall2a
|
mical-like 2a |
| chr15_-_30976483 | 0.73 |
ENSDART00000112511
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr10_+_44456550 | 0.72 |
ENSDART00000157611
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr3_-_40090673 | 0.72 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr25_-_26399777 | 0.71 |
ENSDART00000140643
|
commd4
|
COMM domain containing 4 |
| chr4_+_4825409 | 0.71 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr6_-_39633603 | 0.71 |
ENSDART00000104042
|
atf7b
|
activating transcription factor 7b |
| chr23_-_480433 | 0.71 |
ENSDART00000128433
|
hm13
|
histocompatibility (minor) 13 |
| chr1_-_8801877 | 0.71 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr16_-_52847564 | 0.71 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr16_+_4350448 | 0.70 |
|
|
|
| chr23_+_39961676 | 0.69 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| chr12_+_47729417 | 0.68 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr7_-_73016194 | 0.67 |
|
|
|
| chr25_-_7794186 | 0.67 |
ENSDART00000104686
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr17_-_25630635 | 0.67 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_+_6557967 | 0.67 |
ENSDART00000162255
|
nup85
|
nucleoporin 85 |
| chr14_-_33084418 | 0.67 |
ENSDART00000105688
|
nkap
|
NFKB activating protein |
| chr3_-_54352535 | 0.67 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr2_+_58943872 | 0.67 |
ENSDART00000158860
ENSDART00000067736 |
stk11
|
serine/threonine kinase 11 |
| chr7_+_25554355 | 0.67 |
ENSDART00000149835
|
mtm1
|
myotubularin 1 |
| chr6_-_51771558 | 0.66 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr23_+_38335290 | 0.66 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr9_+_33179226 | 0.65 |
|
|
|
| chr2_-_27892824 | 0.65 |
|
|
|
| chr16_+_25248578 | 0.65 |
ENSDART00000105514
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr20_-_46079529 | 0.65 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr16_-_4350251 | 0.65 |
|
|
|
| chr10_-_33307845 | 0.64 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr17_-_14443647 | 0.64 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8-associated membrane protein |
| chr20_-_22170411 | 0.63 |
ENSDART00000155568
|
BX088688.3
|
ENSDARG00000097598 |
| chr3_-_27159896 | 0.63 |
ENSDART00000151742
|
BX908803.1
|
ENSDARG00000096260 |
| chr14_+_11872906 | 0.63 |
ENSDART00000054626
|
hdac3
|
histone deacetylase 3 |
| chr5_-_71605608 | 0.63 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr11_-_25223594 | 0.63 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr20_+_27188382 | 0.63 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr2_+_55065658 | 0.62 |
|
|
|
| chr22_+_2422443 | 0.62 |
|
|
|
| chr19_+_19806894 | 0.62 |
ENSDART00000170321
|
si:dkey-95j14.1
|
si:dkey-95j14.1 |
| chr16_+_47283374 | 0.61 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr3_+_28729443 | 0.61 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr8_-_19215929 | 0.60 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr6_+_48349630 | 0.60 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr18_+_5199305 | 0.60 |
ENSDART00000020233
|
zdhhc7
|
zinc finger, DHHC-type containing 7 |
| chr5_+_29231012 | 0.60 |
ENSDART00000159327
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr6_-_19556029 | 0.59 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr4_-_14193396 | 0.59 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr3_-_26113336 | 0.59 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr12_+_35101949 | 0.59 |
ENSDART00000153022
|
AL935194.1
|
ENSDARG00000060678 |
| chr17_-_6443673 | 0.58 |
ENSDART00000129950
ENSDART00000154074 |
ANKRD66
|
si:ch211-189e2.2 |
| chr16_-_21064435 | 0.58 |
ENSDART00000040727
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr5_+_69013573 | 0.58 |
ENSDART00000165956
|
setd8b
|
SET domain containing (lysine methyltransferase) 8b |
| chr17_-_7377094 | 0.58 |
|
|
|
| chr11_-_17668381 | 0.58 |
ENSDART00000080752
|
uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr18_+_5199156 | 0.58 |
ENSDART00000020233
|
zdhhc7
|
zinc finger, DHHC-type containing 7 |
| chr24_+_23571680 | 0.57 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr25_-_13394261 | 0.57 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr18_-_7138414 | 0.57 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr11_-_25223110 | 0.56 |
|
|
|
| chr20_-_46079578 | 0.56 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr16_+_26858575 | 0.56 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr17_-_33764220 | 0.56 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr7_-_52850565 | 0.56 |
|
|
|
| chr22_-_10026579 | 0.56 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr5_-_68848113 | 0.55 |
ENSDART00000097249
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr17_-_17739301 | 0.55 |
ENSDART00000157128
|
adck1
|
aarF domain containing kinase 1 |
| chr11_-_18081944 | 0.55 |
ENSDART00000113468
|
CABZ01112215.1
|
ENSDARG00000079534 |
| chr19_-_35286335 | 0.55 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr10_-_14971401 | 0.55 |
ENSDART00000044756
ENSDART00000128579 |
smad2
|
SMAD family member 2 |
| chr23_-_15234716 | 0.55 |
ENSDART00000158943
|
ndrg3b
|
ndrg family member 3b |
| chr8_-_10912137 | 0.54 |
ENSDART00000064058
ENSDART00000162109 ENSDART00000101277 |
pqlc2
|
PQ loop repeat containing 2 |
| chr19_+_1148544 | 0.54 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr6_-_32001100 | 0.54 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr12_-_11311554 | 0.54 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr7_-_25998808 | 0.54 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr7_+_72925910 | 0.54 |
ENSDART00000175267
ENSDART00000179638 |
CABZ01067170.1
|
ENSDARG00000106453 |
| chr15_-_25333252 | 0.54 |
ENSDART00000127771
|
mettl16
|
methyltransferase like 16 |
| chr24_+_23571827 | 0.53 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr8_-_19236074 | 0.53 |
ENSDART00000137994
|
zgc:77486
|
zgc:77486 |
| chr13_+_31415523 | 0.53 |
ENSDART00000076527
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr23_-_27123338 | 0.53 |
ENSDART00000142324
ENSDART00000133249 |
ENSDARG00000025766
|
ENSDARG00000025766 |
| chr2_+_42923652 | 0.53 |
ENSDART00000168318
|
BX005083.1
|
ENSDARG00000097562 |
| chr11_-_25223169 | 0.52 |
|
|
|
| chr16_+_25248338 | 0.52 |
ENSDART00000105514
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr24_+_25871962 | 0.52 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr5_-_53748764 | 0.52 |
ENSDART00000160781
ENSDART00000160965 ENSDART00000161692 |
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr15_-_2647590 | 0.52 |
ENSDART00000157159
|
si:ch211-235m3.10
|
si:ch211-235m3.10 |
| chr8_-_19235896 | 0.52 |
ENSDART00000036148
|
zgc:77486
|
zgc:77486 |
| chr8_+_41003629 | 0.51 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr3_-_36940649 | 0.51 |
ENSDART00000110716
ENSDART00000160233 |
tubg1
|
tubulin, gamma 1 |
| chr7_+_48025010 | 0.51 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr3_-_30357190 | 0.51 |
ENSDART00000055393
|
med25
|
mediator complex subunit 25 |
| chr5_+_29052462 | 0.51 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr24_+_29278310 | 0.51 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr3_+_30927580 | 0.50 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr2_-_32403565 | 0.50 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr4_-_5010148 | 0.50 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr18_-_6896957 | 0.50 |
ENSDART00000175747
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr23_+_19632234 | 0.50 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase, H+ transporting, lysosomal accessory protein 1-like b |
| chr8_-_19437095 | 0.49 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.4 | 2.9 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.4 | 4.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 1.9 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 1.1 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.4 | 1.1 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.3 | 1.4 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.3 | 0.9 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 2.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 0.8 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 0.8 | GO:0002363 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) type 2 immune response(GO:0042092) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T cell selection(GO:0045058) T-helper 2 cell differentiation(GO:0045064) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 1.0 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.2 | 1.0 | GO:0071346 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.2 | 0.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 0.9 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.2 | 1.7 | GO:0048608 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 0.9 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 1.9 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.7 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 1.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 2.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.6 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.1 | 0.4 | GO:0070841 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 2.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0045041 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.4 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.3 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.1 | 2.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.5 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.4 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.5 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0060282 | regulation of oocyte development(GO:0060281) positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.9 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 1.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.5 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 1.4 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 0.6 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.5 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.1 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 0.5 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 1.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.9 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:2000104 | replication fork protection(GO:0048478) negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.6 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 1.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 1.2 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 3.6 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 1.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 5.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 3.8 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 3.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.2 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.2 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.1 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 2.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.0 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 2.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0052907 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 0.4 | 1.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 1.3 | GO:0019705 | protein-cysteine S-myristoyltransferase activity(GO:0019705) |
| 0.3 | 1.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.3 | 1.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 2.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 0.9 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 1.0 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 2.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.2 | 1.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 2.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 1.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 0.5 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.5 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.1 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.2 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.6 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 3.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 2.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 1.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 4.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 2.3 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 1.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.7 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.1 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.3 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |