Project
DANIO-CODE
Navigation
Downloads
Results for barx1
Z-value: 0.68
Transcription factors associated with barx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx1
|
ENSDARG00000007407 | BARX homeobox 1 |
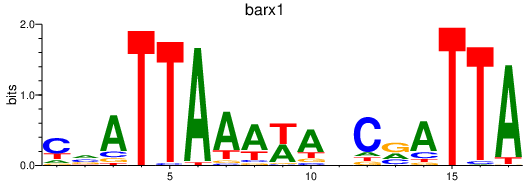
Activity profile of barx1 motif
Sorted Z-values of barx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of barx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_26035166 | 0.77 |
ENSDART00000134939
|
rpl23a
|
ribosomal protein L23a |
| chr5_+_48013258 | 0.70 |
|
|
|
| chr24_-_20782338 | 0.69 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr4_-_14193396 | 0.59 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr1_-_619364 | 0.55 |
ENSDART00000134456
|
atp5j
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_-_49900124 | 0.55 |
ENSDART00000150204
|
atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr13_-_694204 | 0.52 |
ENSDART00000139589
|
polh
|
polymerase (DNA directed), eta |
| chr24_+_20782714 | 0.52 |
ENSDART00000144883
|
fam162a
|
family with sequence similarity 162, member A |
| chr19_-_868378 | 0.51 |
|
|
|
| chr6_-_54172509 | 0.46 |
ENSDART00000149468
|
rps10
|
ribosomal protein S10 |
| chr19_-_10737772 | 0.46 |
ENSDART00000081379
|
olah
|
oleoyl-ACP hydrolase |
| chr7_-_73631286 | 0.46 |
ENSDART00000129254
|
zgc:173552
|
zgc:173552 |
| chr6_-_55342742 | 0.46 |
|
|
|
| chr21_-_5774127 | 0.45 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr1_+_35224410 | 0.45 |
ENSDART00000085021
ENSDART00000140144 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr25_+_34629476 | 0.45 |
ENSDART00000156376
|
CR762436.2
|
Histone H3.2 |
| chr24_+_28571936 | 0.44 |
ENSDART00000153933
|
CR933730.1
|
ENSDARG00000097841 |
| chr13_+_31271568 | 0.44 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr5_-_67218088 | 0.44 |
ENSDART00000166142
|
abhd10a
|
abhydrolase domain containing 10a |
| chr9_+_21495976 | 0.43 |
ENSDART00000004108
ENSDART00000160365 |
xpo4
|
exportin 4 |
| chr10_+_9576840 | 0.42 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| chr2_+_216779 | 0.42 |
ENSDART00000101071
|
zgc:113293
|
zgc:113293 |
| chr23_+_19749426 | 0.42 |
|
|
|
| chr1_-_7968497 | 0.41 |
ENSDART00000141407
ENSDART00000148067 |
actb1
|
actin, beta 1 |
| chr25_+_34629404 | 0.39 |
ENSDART00000156376
|
CR762436.2
|
Histone H3.2 |
| chr4_+_377829 | 0.39 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr11_-_7786415 | 0.39 |
ENSDART00000154569
|
CR450813.1
|
ENSDARG00000097035 |
| chr22_-_18753903 | 0.38 |
ENSDART00000019235
|
atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr11_+_15988149 | 0.38 |
ENSDART00000168083
|
Metazoa_SRP
|
Metazoan signal recognition particle RNA |
| chr19_+_21193691 | 0.38 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr7_+_58866578 | 0.37 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr14_-_10081301 | 0.37 |
ENSDART00000165101
|
commd5
|
COMM domain containing 5 |
| chr22_+_26616977 | 0.36 |
ENSDART00000171972
ENSDART00000083351 |
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr25_+_34559650 | 0.36 |
ENSDART00000073454
|
zgc:110434
|
zgc:110434 |
| chr23_-_37010705 | 0.36 |
ENSDART00000134461
|
zgc:193690
|
zgc:193690 |
| chr14_+_24637864 | 0.35 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr19_-_11397220 | 0.34 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr19_-_48711616 | 0.34 |
ENSDART00000169464
|
rpl14
|
ribosomal protein L14 |
| chr3_-_29768364 | 0.33 |
ENSDART00000020311
|
rpl27
|
ribosomal protein L27 |
| chr6_-_40771121 | 0.32 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr3_+_28371725 | 0.32 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr21_+_10152870 | 0.32 |
|
|
|
| chr1_+_30337385 | 0.32 |
ENSDART00000148229
|
BX284697.1
|
ENSDARG00000093358 |
| chr4_-_15005050 | 0.32 |
ENSDART00000067048
|
klhdc10
|
kelch domain containing 10 |
| chr17_-_45036982 | 0.31 |
ENSDART00000155043
|
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr11_+_30497103 | 0.31 |
ENSDART00000132491
|
wdr83os
|
WD repeat domain 83 opposite strand |
| chr3_+_59589031 | 0.31 |
ENSDART00000108647
|
alyref
|
Aly/REF export factor |
| chr21_+_5027668 | 0.31 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr3_-_61122514 | 0.31 |
|
|
|
| chr2_+_3671135 | 0.31 |
ENSDART00000113395
|
SERP1
|
stress associated endoplasmic reticulum protein 1 |
| chr15_+_855739 | 0.31 |
ENSDART00000046783
|
zgc:113294
|
zgc:113294 |
| chr10_+_9576710 | 0.30 |
ENSDART00000080843
|
ndufa8
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 |
| KN150123v1_-_37214 | 0.30 |
|
|
|
| chr2_+_54911012 | 0.30 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr11_-_16017396 | 0.29 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr6_-_1638559 | 0.29 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr25_-_11287103 | 0.29 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr19_+_33267939 | 0.29 |
ENSDART00000176726
|
rpl30
|
ribosomal protein L30 |
| chr5_-_61279732 | 0.29 |
ENSDART00000097338
|
napaa
|
N-ethylmaleimide-sensitive factor attachment protein, alpha a |
| chr17_-_7635061 | 0.28 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr4_-_71830240 | 0.28 |
|
|
|
| chr15_+_44177774 | 0.28 |
|
|
|
| chr3_-_61144538 | 0.28 |
ENSDART00000155932
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr5_-_11779717 | 0.27 |
ENSDART00000143646
|
BX276096.1
|
ENSDARG00000093726 |
| chr5_-_4483970 | 0.27 |
|
|
|
| chr12_+_33874082 | 0.27 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr9_-_55126838 | 0.27 |
ENSDART00000162158
ENSDART00000168430 |
trappc2
|
trafficking protein particle complex 2 |
| chr17_+_12788580 | 0.26 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr24_+_20772309 | 0.25 |
ENSDART00000043193
ENSDART00000136663 |
cst14a.1
|
cystatin 14a, tandem duplicate 1 |
| chr21_-_27164751 | 0.25 |
ENSDART00000065402
|
zgc:77262
|
zgc:77262 |
| chr2_-_41269732 | 0.24 |
ENSDART00000134756
|
hs6st1a
|
heparan sulfate 6-O-sulfotransferase 1a |
| chr16_+_38209664 | 0.24 |
ENSDART00000160332
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr23_+_41129663 | 0.24 |
|
|
|
| KN150703v1_-_32804 | 0.24 |
|
|
|
| chr3_+_32617947 | 0.24 |
ENSDART00000125260
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr3_-_32205873 | 0.23 |
ENSDART00000156918
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr19_-_27093095 | 0.23 |
|
|
|
| chr7_+_15059782 | 0.23 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr25_+_20021806 | 0.23 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr5_-_4447263 | 0.23 |
|
|
|
| chr23_-_40873523 | 0.23 |
ENSDART00000127420
|
ENSDARG00000088040
|
ENSDARG00000088040 |
| chr19_-_27093002 | 0.23 |
|
|
|
| chr5_-_39077051 | 0.23 |
|
|
|
| chr19_-_31996936 | 0.23 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr1_+_9176411 | 0.23 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr25_+_32716795 | 0.23 |
ENSDART00000131098
|
ENSDARG00000087402
|
ENSDARG00000087402 |
| chr5_-_53907908 | 0.22 |
ENSDART00000158069
|
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr3_+_27667194 | 0.22 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr10_+_20632757 | 0.22 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr21_+_16969092 | 0.22 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr20_+_54512601 | 0.22 |
ENSDART00000169386
|
faua
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a |
| chr5_-_23211957 | 0.22 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr25_-_34631252 | 0.22 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr14_-_30630835 | 0.22 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chr15_-_26638024 | 0.21 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr17_-_7061096 | 0.21 |
|
|
|
| chr1_-_51870424 | 0.21 |
ENSDART00000135636
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr14_-_16171165 | 0.21 |
ENSDART00000089021
|
canx
|
calnexin |
| chr23_+_31986806 | 0.21 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr3_+_12087549 | 0.21 |
|
|
|
| chr21_-_40694583 | 0.21 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr6_+_36862078 | 0.21 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr1_+_38099962 | 0.21 |
ENSDART00000166864
|
spcs3
|
signal peptidase complex subunit 3 |
| chr24_+_12845339 | 0.21 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr20_-_46630367 | 0.21 |
ENSDART00000060689
|
eif2b2
|
eukaryotic translation initiation factor 2B, subunit 2 beta |
| chr22_-_10026610 | 0.21 |
ENSDART00000179409
|
si:ch211-222k6.1
|
si:ch211-222k6.1 |
| chr14_-_46525518 | 0.21 |
ENSDART00000173172
|
si:ch211-114c17.1
|
si:ch211-114c17.1 |
| chr24_+_12839000 | 0.21 |
ENSDART00000145214
|
tceb1a
|
transcription elongation factor B (SIII), polypeptide 1a |
| chr25_+_19642595 | 0.21 |
|
|
|
| chr14_+_159572 | 0.20 |
|
|
|
| chr23_-_294139 | 0.20 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr5_-_48013093 | 0.20 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr13_+_4239960 | 0.20 |
ENSDART00000135113
|
sft2d1
|
SFT2 domain containing 1 |
| chr3_+_25776714 | 0.20 |
ENSDART00000170324
|
tom1
|
target of myb1 membrane trafficking protein |
| chr9_-_15453604 | 0.20 |
ENSDART00000124346
ENSDART00000010521 ENSDART00000103753 |
fn1a
|
fibronectin 1a |
| chr9_-_41238670 | 0.20 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr17_-_24802344 | 0.20 |
ENSDART00000156606
|
trmt61b
|
tRNA methyltransferase 61B |
| chr17_-_30635487 | 0.20 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr10_+_3052919 | 0.20 |
ENSDART00000028870
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr25_+_18997277 | 0.20 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr14_-_10081393 | 0.20 |
ENSDART00000165101
|
commd5
|
COMM domain containing 5 |
| chr22_+_2014357 | 0.20 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr15_+_44177693 | 0.20 |
|
|
|
| chr24_-_6048914 | 0.20 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr19_+_32614657 | 0.19 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr6_-_2501966 | 0.19 |
ENSDART00000155109
|
hyi
|
hydroxypyruvate isomerase |
| chr14_+_45863932 | 0.19 |
ENSDART00000074216
|
higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr1_+_51087450 | 0.19 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr21_+_27152225 | 0.19 |
ENSDART00000125349
|
bada
|
BCL2-associated agonist of cell death a |
| chr3_+_15743134 | 0.19 |
ENSDART00000114564
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr8_+_50953220 | 0.19 |
ENSDART00000124748
|
b2ml
|
beta-2-microglobulin, like |
| chr15_-_25025058 | 0.19 |
ENSDART00000109990
|
abhd15a
|
abhydrolase domain containing 15a |
| chr22_+_1268270 | 0.19 |
ENSDART00000171750
|
si:ch73-138e16.3
|
si:ch73-138e16.3 |
| chr15_-_743589 | 0.19 |
ENSDART00000156200
|
zgc:174574
|
zgc:174574 |
| chr22_-_510040 | 0.19 |
ENSDART00000140101
|
ccnd3
|
cyclin D3 |
| chr3_+_18786265 | 0.19 |
ENSDART00000141637
|
si:ch211-198m1.1
|
si:ch211-198m1.1 |
| chr2_-_39049391 | 0.18 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr3_-_61154674 | 0.18 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr23_+_25953162 | 0.18 |
ENSDART00000138731
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr4_+_377994 | 0.18 |
ENSDART00000148933
|
rpl18a
|
ribosomal protein L18a |
| chr9_+_52256434 | 0.18 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr7_-_6231474 | 0.18 |
ENSDART00000159524
|
CU457819.1
|
Histone H3.2 |
| KN150266v1_+_74105 | 0.18 |
|
|
|
| chr1_+_29027527 | 0.18 |
ENSDART00000142184
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr7_-_6222134 | 0.18 |
ENSDART00000173197
|
hist1h4l
|
histone 1, H4, like |
| chr10_+_10354768 | 0.18 |
ENSDART00000142895
|
urm1
|
ubiquitin related modifier 1 |
| chr17_+_24595050 | 0.18 |
ENSDART00000064738
|
atpif1b
|
ATPase inhibitory factor 1b |
| chr3_-_40791035 | 0.18 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr1_+_52739724 | 0.18 |
ENSDART00000159220
|
ucp1
|
uncoupling protein 1 |
| chr9_-_31467299 | 0.18 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr11_+_328781 | 0.18 |
ENSDART00000147426
|
pdrg1
|
p53 and DNA-damage regulated 1 |
| chr24_+_3296536 | 0.18 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr25_+_28450398 | 0.18 |
ENSDART00000075151
|
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr10_-_42302932 | 0.18 |
ENSDART00000076693
ENSDART00000073631 |
stambpa
|
STAM binding protein a |
| chr4_-_6407602 | 0.18 |
ENSDART00000134376
|
CR356233.2
|
ENSDARG00000093020 |
| chr24_+_16760759 | 0.18 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr13_+_42276937 | 0.18 |
|
|
|
| chr21_-_45169199 | 0.17 |
ENSDART00000149766
|
vdac1
|
voltage-dependent anion channel 1 |
| chr12_-_29928951 | 0.17 |
ENSDART00000149077
ENSDART00000148847 |
trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr6_+_247024 | 0.17 |
ENSDART00000123769
|
rps19bp1
|
ribosomal protein S19 binding protein 1 |
| chr23_-_24299965 | 0.17 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr19_+_5052459 | 0.17 |
ENSDART00000003634
|
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr9_+_45193290 | 0.17 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr9_+_24398747 | 0.17 |
ENSDART00000090280
ENSDART00000133479 |
zgc:153901
|
zgc:153901 |
| chr3_-_3646681 | 0.17 |
ENSDART00000165648
ENSDART00000046454 ENSDART00000115282 ENSDART00000164463 |
ENSDARG00000076771
|
ENSDARG00000076771 |
| chr1_+_518777 | 0.17 |
ENSDART00000109083
|
txnl4b
|
thioredoxin-like 4B |
| chr7_-_71214600 | 0.17 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr18_+_3475225 | 0.17 |
ENSDART00000169814
|
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr21_+_30625592 | 0.17 |
ENSDART00000138664
|
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr3_+_24510974 | 0.16 |
ENSDART00000148414
ENSDART00000055590 |
zgc:113411
|
zgc:113411 |
| chr17_-_20675997 | 0.16 |
ENSDART00000063613
ENSDART00000171575 |
ccdc6a
|
coiled-coil domain containing 6a |
| chr19_-_45209699 | 0.16 |
|
|
|
| chr11_+_42433566 | 0.16 |
ENSDART00000162327
|
zgc:194981
|
zgc:194981 |
| chr5_-_10966029 | 0.16 |
|
|
|
| chr25_-_18338974 | 0.16 |
ENSDART00000004771
|
poc1b
|
POC1 centriolar protein B |
| chr23_+_16848278 | 0.16 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr8_+_44619220 | 0.16 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr25_-_34596585 | 0.16 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr5_+_12028418 | 0.16 |
ENSDART00000131644
|
rfc5
|
replication factor C (activator 1) 5 |
| chr6_-_9933233 | 0.16 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr15_-_19396871 | 0.16 |
|
|
|
| chr23_+_30908755 | 0.16 |
ENSDART00000154145
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr25_-_34596670 | 0.16 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr20_+_13987346 | 0.16 |
ENSDART00000152611
|
nek2
|
NIMA-related kinase 2 |
| chr4_-_12478339 | 0.16 |
ENSDART00000027756
|
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr12_+_36414886 | 0.16 |
ENSDART00000171268
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_55195428 | 0.16 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr5_-_9179162 | 0.16 |
|
|
|
| chr5_-_48013023 | 0.16 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr2_-_37479961 | 0.16 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr5_-_67378805 | 0.16 |
ENSDART00000177026
|
rnf167
|
ring finger protein 167 |
| chr8_-_44617324 | 0.16 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr3_-_40790889 | 0.16 |
|
|
|
| chr20_-_18895415 | 0.16 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr5_+_4906690 | 0.15 |
ENSDART00000124022
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr9_-_42682501 | 0.15 |
ENSDART00000048320
|
tfpia
|
tissue factor pathway inhibitor a |
| chr1_-_50576110 | 0.15 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr16_+_12726556 | 0.15 |
|
|
|
| chr1_+_43471879 | 0.15 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_-_55195566 | 0.15 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0090171 | chondrocyte morphogenesis(GO:0090171) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0032447 | protein urmylation(GO:0032447) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.0 | 0.1 | GO:0071031 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:2000040 | planar cell polarity pathway involved in axis elongation(GO:0003402) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) |
| 0.0 | 0.1 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0010984 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0046477 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.0 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.0 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.0 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.0 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 2.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 1.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.0 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.1 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0042923 | opioid receptor activity(GO:0004985) neuropeptide binding(GO:0042923) |
| 0.1 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.1 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.1 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.0 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.0 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 3.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |