Project
DANIO-CODE
Navigation
Downloads
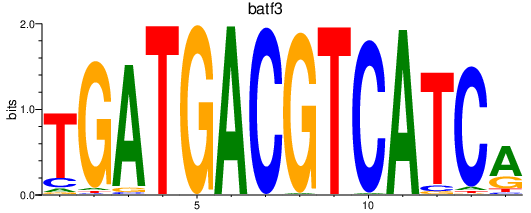
Results for batf3
Z-value: 1.53
Transcription factors associated with batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
batf3
|
ENSDARG00000042577 | basic leucine zipper transcription factor, ATF-like 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| batf3 | dr10_dc_chr20_-_37910887_37910994 | 0.77 | 5.0e-04 | Click! |
Activity profile of batf3 motif
Sorted Z-values of batf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of batf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_27680365 | 5.74 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr23_-_1562572 | 5.47 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr22_+_1500412 | 5.37 |
ENSDART00000160406
|
si:ch211-255f4.5
|
si:ch211-255f4.5 |
| chr22_+_1837102 | 5.18 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr4_-_11126782 | 4.93 |
ENSDART00000150419
|
BX784029.1
|
ENSDARG00000096039 |
| chr22_+_2014357 | 3.87 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr22_+_2014149 | 3.76 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr25_-_6320310 | 3.51 |
ENSDART00000139606
|
snupn
|
snurportin 1 |
| chr15_+_35105420 | 3.30 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr16_-_29452509 | 3.17 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr17_+_15666078 | 3.10 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr22_+_1905394 | 2.99 |
ENSDART00000165758
|
znf1181
|
zinc finger protein 1181 |
| chr22_+_1445227 | 2.91 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr3_-_16263615 | 2.71 |
ENSDART00000127309
|
eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr11_+_24076334 | 2.71 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr17_+_15666420 | 2.63 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr22_+_1462211 | 2.55 |
|
|
|
| chr22_+_1894340 | 2.54 |
ENSDART00000164158
|
znf1156
|
zinc finger protein 1156 |
| chr22_+_1484719 | 2.43 |
ENSDART00000159973
|
zgc:173486
|
zgc:173486 |
| chr22_+_2422443 | 2.37 |
|
|
|
| chr22_+_1451958 | 2.31 |
|
|
|
| chr1_+_9621488 | 2.30 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr17_-_14583728 | 2.11 |
ENSDART00000174703
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr11_+_27919002 | 2.10 |
|
|
|
| chr6_-_1685041 | 2.08 |
|
|
|
| chr5_-_27953580 | 2.00 |
ENSDART00000135127
|
zgc:162952
|
zgc:162952 |
| chr20_-_20921480 | 1.97 |
ENSDART00000152781
|
ckbb
|
creatine kinase, brain b |
| chr22_+_2386743 | 1.88 |
ENSDART00000132925
|
zgc:112977
|
zgc:112977 |
| chr1_+_9621612 | 1.85 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr22_+_1445302 | 1.78 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr22_+_2014232 | 1.78 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr16_+_53391438 | 1.76 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr23_-_1562603 | 1.71 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr16_-_41716834 | 1.71 |
ENSDART00000084528
|
atp2c1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_+_1624585 | 1.66 |
ENSDART00000159804
|
si:dkey-1b17.2
|
si:dkey-1b17.2 |
| chr5_-_27953622 | 1.56 |
ENSDART00000135127
|
zgc:162952
|
zgc:162952 |
| chr22_+_2785213 | 1.55 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr11_+_24076371 | 1.51 |
ENSDART00000166045
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_-_18074946 | 1.46 |
|
|
|
| chr22_+_2644250 | 1.44 |
ENSDART00000031485
|
znf1162
|
zinc finger protein 1162 |
| chr22_+_2231377 | 1.32 |
ENSDART00000143366
|
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr5_-_27953681 | 1.30 |
ENSDART00000078708
|
zgc:162952
|
zgc:162952 |
| chr14_-_32619461 | 1.25 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr16_+_53391776 | 1.20 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr7_-_20378231 | 1.08 |
|
|
|
| chr21_-_14666277 | 0.92 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| KN149782v1_+_35344 | 0.83 |
|
|
|
| chr7_-_20378183 | 0.80 |
|
|
|
| chr22_+_1540035 | 0.79 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr13_+_40628694 | 0.74 |
ENSDART00000143051
|
BX294129.1
|
ENSDARG00000092014 |
| chr22_+_1923642 | 0.64 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr7_+_8424495 | 0.60 |
ENSDART00000172511
|
si:ch211-1o7.2
|
si:ch211-1o7.2 |
| chr21_-_14666223 | 0.39 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr22_+_2166859 | 0.30 |
ENSDART00000159279
|
znf1152
|
zinc finger protein 1152 |
| KN149782v1_-_35118 | 0.29 |
|
|
|
| chr3_-_40020899 | 0.28 |
ENSDART00000025285
|
drg2
|
developmentally regulated GTP binding protein 2 |
| chr22_+_2074020 | 0.26 |
ENSDART00000106540
|
znf1161
|
zinc finger protein 1161 |
| chr6_-_13881001 | 0.15 |
ENSDART00000004656
|
ENSDARG00000020187
|
ENSDARG00000020187 |
| chr22_+_1691850 | 0.10 |
ENSDART00000170471
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr23_-_40873523 | 0.09 |
ENSDART00000127420
|
ENSDARG00000088040
|
ENSDARG00000088040 |
| chr5_+_11311820 | 0.09 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr22_+_30381331 | 0.09 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr5_-_11312085 | 0.08 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.8 | 4.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 3.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 2.0 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 4.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.1 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 3.2 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 4.9 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 36.9 | GO:0006357 | regulation of transcription from RNA polymerase II promoter(GO:0006357) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 2.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 6.1 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 1.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 4.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 2.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 3.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 4.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 4.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 7.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 34.6 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 4.9 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |