Project
DANIO-CODE
Navigation
Downloads
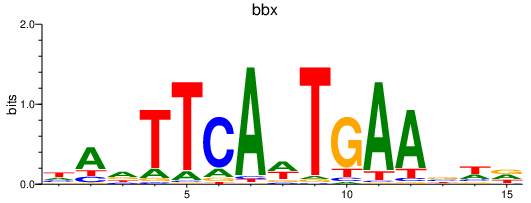
Results for bbx
Z-value: 0.85
Transcription factors associated with bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bbx
|
ENSDARG00000012699 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bbx | dr10_dc_chr10_-_28638101_28638195 | -0.57 | 2.1e-02 | Click! |
Activity profile of bbx motif
Sorted Z-values of bbx motif
Network of associatons between targets according to the STRING database.
First level regulatory network of bbx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_18446876 | 1.59 |
ENSDART00000039693
|
pgp
|
phosphoglycolate phosphatase |
| chr6_+_36862078 | 0.99 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr3_+_32657833 | 0.80 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr12_+_23691261 | 0.72 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr15_+_44177774 | 0.71 |
|
|
|
| chr15_+_44177693 | 0.71 |
|
|
|
| chr11_-_2385431 | 0.68 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr24_-_20782338 | 0.65 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr5_-_55263153 | 0.59 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr6_+_36861678 | 0.59 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr8_-_51380776 | 0.58 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr17_+_24421150 | 0.53 |
ENSDART00000168926
|
mdh1ab
|
malate dehydrogenase 1Ab, NAD (soluble) |
| chr12_-_18446766 | 0.53 |
ENSDART00000039693
|
pgp
|
phosphoglycolate phosphatase |
| chr19_-_3833037 | 0.53 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr15_-_20254259 | 0.52 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr25_+_20118286 | 0.51 |
ENSDART00000104297
|
tnnt2d
|
troponin T2d, cardiac |
| chr3_+_12087549 | 0.48 |
|
|
|
| chr19_+_43745687 | 0.48 |
ENSDART00000102384
|
sesn2
|
sestrin 2 |
| chr15_-_35354323 | 0.47 |
ENSDART00000144944
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr16_+_54579976 | 0.46 |
|
|
|
| chr21_+_21227307 | 0.45 |
ENSDART00000159110
ENSDART00000089651 |
ccdc61
|
coiled-coil domain containing 61 |
| chr15_-_35354406 | 0.45 |
ENSDART00000124780
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr11_+_8142982 | 0.44 |
ENSDART00000166379
|
samd13
|
sterile alpha motif domain containing 13 |
| chr1_+_35224410 | 0.44 |
ENSDART00000085021
ENSDART00000140144 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_-_23003738 | 0.44 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr23_+_22729939 | 0.43 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr23_+_35405877 | 0.43 |
ENSDART00000159218
|
acp1
|
acid phosphatase 1, soluble |
| chr14_+_33117964 | 0.42 |
ENSDART00000075278
|
atp1b4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr19_-_3832999 | 0.37 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr23_-_18454945 | 0.37 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr9_+_19812730 | 0.37 |
ENSDART00000110009
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr7_+_50123055 | 0.36 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr17_+_23529112 | 0.36 |
ENSDART00000063953
|
zgc:65997
|
zgc:65997 |
| chr5_+_36049836 | 0.36 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr10_-_14582293 | 0.36 |
ENSDART00000039161
|
hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr5_-_21523599 | 0.36 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr5_-_47106694 | 0.36 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr18_+_38422567 | 0.36 |
ENSDART00000178559
ENSDART00000175878 |
CU856178.1
|
ENSDARG00000108097 |
| chr8_-_37023410 | 0.35 |
ENSDART00000111513
|
si:ch211-218o21.4
|
si:ch211-218o21.4 |
| chr15_-_16047125 | 0.35 |
ENSDART00000166583
|
synrg
|
synergin, gamma |
| chr6_-_8075384 | 0.35 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr23_+_3778116 | 0.34 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr23_+_3778066 | 0.34 |
ENSDART00000141880
|
smim29
|
small integral membrane protein 29 |
| chr3_-_14545005 | 0.34 |
ENSDART00000133850
|
gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr2_-_23003710 | 0.33 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr6_-_6835861 | 0.33 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr9_+_19812539 | 0.32 |
ENSDART00000147662
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr24_-_42053682 | 0.32 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr16_+_32060521 | 0.32 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr5_+_37144525 | 0.32 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr14_-_4070037 | 0.32 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr3_+_54507075 | 0.31 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr6_+_9717087 | 0.30 |
ENSDART00000019325
|
cyp20a1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr2_-_41669008 | 0.30 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr7_+_58541247 | 0.30 |
ENSDART00000165852
|
lypla1
|
lysophospholipase I |
| chr21_+_13689239 | 0.30 |
ENSDART00000112451
|
slc31a2
|
solute carrier family 31 (copper transporter), member 2 |
| chr19_+_43745546 | 0.28 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr2_+_23004159 | 0.28 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr8_-_30311299 | 0.27 |
|
|
|
| chr23_-_39367876 | 0.27 |
|
|
|
| chr23_-_3778284 | 0.27 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr11_-_22200590 | 0.26 |
ENSDART00000006580
|
tfeb
|
transcription factor EB |
| chr8_+_21082244 | 0.25 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| KN149895v1_+_73008 | 0.25 |
|
|
|
| chr1_+_18842158 | 0.25 |
ENSDART00000103089
|
ENSDARG00000070320
|
ENSDARG00000070320 |
| chr11_-_27253835 | 0.25 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr13_+_1595986 | 0.25 |
|
|
|
| chr19_+_33506239 | 0.24 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr9_-_24309717 | 0.24 |
ENSDART00000140808
|
tyw5
|
tRNA-yW synthesizing protein 5 |
| chr20_+_51034244 | 0.24 |
|
|
|
| chr5_+_60871396 | 0.23 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr25_-_12633686 | 0.23 |
ENSDART00000171144
ENSDART00000171834 |
klhdc4
|
kelch domain containing 4 |
| chr23_-_24416864 | 0.23 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr8_+_26037704 | 0.22 |
|
|
|
| chr2_-_30005139 | 0.22 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr15_-_16950049 | 0.21 |
|
|
|
| chr9_-_48709908 | 0.21 |
ENSDART00000058248
|
ENSDARG00000039824
|
ENSDARG00000039824 |
| chr4_+_12033088 | 0.20 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr13_+_30968221 | 0.20 |
ENSDART00000014922
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr7_+_36627685 | 0.20 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr8_-_51380846 | 0.20 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr6_+_36862148 | 0.20 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr13_-_22964527 | 0.20 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr16_-_45269179 | 0.20 |
ENSDART00000162095
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr15_-_12297979 | 0.19 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr7_+_5788679 | 0.19 |
ENSDART00000173232
|
CU459186.7
|
ENSDARG00000105397 |
| chr9_+_56182756 | 0.18 |
ENSDART00000144757
|
mxra5b
|
matrix-remodelling associated 5b |
| chr4_-_71906883 | 0.18 |
ENSDART00000150753
|
znf1015
|
zinc finger protein 1015 |
| chr24_-_16760393 | 0.18 |
ENSDART00000149706
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr6_+_14854101 | 0.18 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr25_-_5776878 | 0.18 |
ENSDART00000178323
|
CABZ01067754.1
|
ENSDARG00000107684 |
| chr17_+_22359660 | 0.18 |
ENSDART00000162670
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr13_+_18376540 | 0.18 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr13_-_24538905 | 0.18 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr2_-_31319228 | 0.18 |
ENSDART00000034784
|
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr23_-_24416911 | 0.18 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr3_-_14545180 | 0.17 |
ENSDART00000133850
|
gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr24_-_8861608 | 0.16 |
ENSDART00000150185
|
mppe1
|
metallophosphoesterase 1 |
| chr3_+_31047152 | 0.16 |
|
|
|
| chr7_+_15629256 | 0.15 |
ENSDART00000138689
ENSDART00000137235 |
pax6b
|
paired box 6b |
| chr20_-_39833314 | 0.15 |
ENSDART00000101049
|
tpd52l1
|
tumor protein D52-like 1 |
| chr8_-_33145412 | 0.15 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr8_+_20406797 | 0.14 |
ENSDART00000016422
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr16_-_32059848 | 0.13 |
ENSDART00000138701
|
gstk1
|
glutathione S-transferase kappa 1 |
| chr16_+_25401922 | 0.13 |
ENSDART00000172514
ENSDART00000086409 |
dync1i1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr17_+_44583892 | 0.13 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr12_+_4186084 | 0.13 |
ENSDART00000144881
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr16_-_41515399 | 0.12 |
ENSDART00000169116
|
cpne4a
|
copine IVa |
| chr15_+_16950144 | 0.12 |
ENSDART00000049196
|
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr22_+_21221777 | 0.11 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr22_+_21221950 | 0.11 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr8_+_3373066 | 0.11 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr4_+_9668755 | 0.10 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr24_-_12836986 | 0.10 |
ENSDART00000133166
|
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr13_+_24538458 | 0.10 |
|
|
|
| chr9_-_34145854 | 0.10 |
|
|
|
| chr3_-_12087518 | 0.10 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr1_+_21037251 | 0.09 |
|
|
|
| chr7_+_73583860 | 0.09 |
ENSDART00000166244
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr23_+_19743720 | 0.09 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr21_+_13236509 | 0.08 |
|
|
|
| chr11_-_16067646 | 0.08 |
|
|
|
| chr12_-_17564384 | 0.08 |
ENSDART00000079065
|
ccz1
|
CCZ1 homolog, vacuolar protein trafficking and biogenesis associated |
| chr11_-_12121214 | 0.08 |
ENSDART00000133236
ENSDART00000104226 |
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr6_-_39346614 | 0.08 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr21_+_29040831 | 0.08 |
ENSDART00000166403
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr7_+_10460159 | 0.06 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr7_+_36627787 | 0.06 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr25_-_6096124 | 0.06 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr16_+_17069742 | 0.06 |
ENSDART00000132150
|
rpl18
|
ribosomal protein L18 |
| chr12_-_45897929 | 0.06 |
|
|
|
| chr9_-_1949904 | 0.05 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr20_-_25727145 | 0.05 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr12_-_31369772 | 0.05 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr18_+_6898118 | 0.05 |
ENSDART00000018735
|
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr11_-_12121242 | 0.04 |
ENSDART00000136108
|
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr16_-_24280209 | 0.04 |
ENSDART00000048599
|
rps19
|
ribosomal protein S19 |
| chr23_-_3778250 | 0.04 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr16_+_32060609 | 0.04 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr16_+_12938830 | 0.04 |
|
|
|
| chr9_-_15453604 | 0.03 |
ENSDART00000124346
ENSDART00000010521 ENSDART00000103753 |
fn1a
|
fibronectin 1a |
| chr13_-_24538832 | 0.03 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr23_+_44951868 | 0.02 |
|
|
|
| chr14_+_35906366 | 0.02 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr17_+_8642373 | 0.02 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr10_-_13549869 | 0.00 |
|
|
|
| chr16_+_14817799 | 0.00 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.7 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.8 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.2 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.3 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.5 | GO:0098534 | centriole assembly(GO:0098534) |
| 0.0 | 0.3 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 1.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.7 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |