Project
DANIO-CODE
Navigation
Downloads
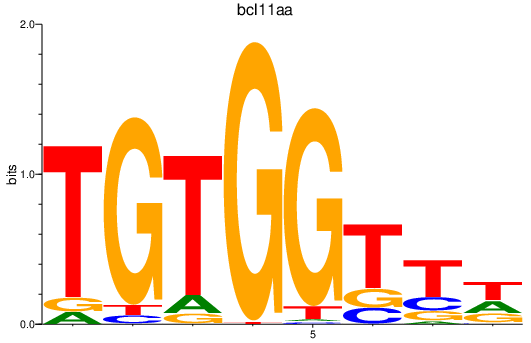
Results for bcl11aa
Z-value: 0.82
Transcription factors associated with bcl11aa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl11aa
|
ENSDARG00000061352 | BAF chromatin remodeling complex subunit BCL11A a |
Activity profile of bcl11aa motif
Sorted Z-values of bcl11aa motif
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl11aa
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_40364675 | 1.85 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr2_+_4366169 | 1.61 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr20_+_34640226 | 1.36 |
ENSDART00000076946
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr19_+_14197118 | 1.34 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr7_-_33994315 | 1.33 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr2_+_36916494 | 1.31 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr24_+_19374200 | 1.26 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr8_+_47197648 | 1.23 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr15_-_26703761 | 1.22 |
ENSDART00000087632
|
ENSDARG00000079340
|
ENSDARG00000079340 |
| KN149817v1_+_2207 | 1.14 |
|
|
|
| chr19_+_30210743 | 1.07 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr6_+_30681170 | 1.05 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr11_+_5467173 | 1.05 |
|
|
|
| chr14_+_24548729 | 1.03 |
|
|
|
| chr2_+_4366348 | 0.98 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr24_+_19374099 | 0.94 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr21_-_11539798 | 0.83 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr11_+_7570027 | 0.79 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr9_-_14533551 | 0.75 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr11_+_7570097 | 0.74 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr6_-_27963880 | 0.72 |
ENSDART00000163747
|
BX914220.1
|
ENSDARG00000098603 |
| chr25_-_22791839 | 0.71 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr14_+_38445969 | 0.68 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr8_+_47197712 | 0.66 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr17_-_26849495 | 0.64 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr2_+_4366022 | 0.62 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr15_-_34709255 | 0.60 |
ENSDART00000099792
|
sostdc1a
|
sclerostin domain containing 1a |
| chr8_-_31359923 | 0.60 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr13_-_5953330 | 0.59 |
ENSDART00000099224
|
dld
|
deltaD |
| chr6_+_45917081 | 0.57 |
ENSDART00000149450
ENSDART00000149642 |
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr25_-_22791921 | 0.54 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr5_-_34940744 | 0.53 |
ENSDART00000145779
|
si:dkey-204l11.1
|
si:dkey-204l11.1 |
| chr14_+_21725253 | 0.51 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr5_+_34022151 | 0.50 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr18_+_22285328 | 0.49 |
|
|
|
| chr5_-_31301280 | 0.44 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr22_-_20284307 | 0.44 |
ENSDART00000159604
|
mbd3b
|
methyl-CpG binding domain protein 3b |
| chr6_-_7281412 | 0.44 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr3_-_25239180 | 0.37 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr23_+_39227734 | 0.36 |
ENSDART00000065331
|
sall4
|
spalt-like transcription factor 4 |
| chr19_+_30210491 | 0.36 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr4_+_32779187 | 0.34 |
ENSDART00000172413
|
znf1042
|
zinc finger protein 1042 |
| chr12_+_18622682 | 0.34 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr11_+_7570326 | 0.34 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr9_-_12726136 | 0.34 |
|
|
|
| chr2_+_4366568 | 0.34 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr10_+_38582701 | 0.33 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr23_+_36208765 | 0.32 |
ENSDART00000127174
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr2_-_24661477 | 0.31 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr15_+_29729531 | 0.30 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr19_-_10996809 | 0.28 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr24_-_21324086 | 0.27 |
|
|
|
| chr2_+_4366515 | 0.27 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr1_-_10217300 | 0.26 |
|
|
|
| chr7_-_25991743 | 0.26 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr11_-_18068313 | 0.26 |
|
|
|
| chr13_+_24703728 | 0.25 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr19_+_14197020 | 0.25 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr6_-_52714750 | 0.25 |
|
|
|
| chr20_+_34640189 | 0.25 |
ENSDART00000076946
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr22_+_9841541 | 0.25 |
ENSDART00000141085
|
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr19_+_30211052 | 0.24 |
ENSDART00000051804
|
marcksl1b
|
MARCKS-like 1b |
| chr23_+_41015697 | 0.23 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr11_+_24582928 | 0.22 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr25_-_19951695 | 0.21 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr18_+_6692267 | 0.20 |
|
|
|
| chr11_+_8119829 | 0.20 |
ENSDART00000011183
|
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr19_+_31046291 | 0.20 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr23_-_39227643 | 0.18 |
|
|
|
| chr24_+_37834148 | 0.18 |
|
|
|
| chr13_+_24703802 | 0.18 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr3_-_6002591 | 0.17 |
|
|
|
| chr3_-_25238896 | 0.16 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr6_-_40654280 | 0.16 |
|
|
|
| chr19_+_10640023 | 0.16 |
ENSDART00000148353
|
usf2
|
upstream transcription factor 2, c-fos interacting |
| chr25_+_15901561 | 0.15 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr16_+_26566014 | 0.15 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr6_-_7281346 | 0.13 |
ENSDART00000133096
|
fkbp11
|
FK506 binding protein 11 |
| chr25_+_5232865 | 0.13 |
|
|
|
| chr19_-_22904687 | 0.12 |
ENSDART00000141503
|
pleca
|
plectin a |
| chr23_+_36208816 | 0.12 |
ENSDART00000127174
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr21_-_10698972 | 0.11 |
|
|
|
| chr4_+_18975027 | 0.11 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr16_+_25425398 | 0.10 |
|
|
|
| chr2_+_12472224 | 0.10 |
ENSDART00000013454
|
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr19_+_30211126 | 0.09 |
ENSDART00000051804
|
marcksl1b
|
MARCKS-like 1b |
| chr23_-_17174825 | 0.09 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr2_+_22196971 | 0.08 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr8_+_26336302 | 0.08 |
|
|
|
| chr15_+_40130393 | 0.08 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr25_+_5232982 | 0.08 |
|
|
|
| chr3_+_26603352 | 0.08 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr9_+_30301667 | 0.08 |
ENSDART00000013591
|
tfg
|
trk-fused gene |
| chr23_-_3813451 | 0.08 |
|
|
|
| chr10_-_22180856 | 0.08 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr4_+_16721264 | 0.08 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr13_+_18130206 | 0.08 |
|
|
|
| chr5_-_21578927 | 0.07 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr3_+_54506912 | 0.07 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr20_+_15119519 | 0.07 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr7_-_25991876 | 0.06 |
ENSDART00000101109
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr2_+_43693486 | 0.06 |
|
|
|
| chr19_-_11480368 | 0.06 |
ENSDART00000080762
|
sept7a
|
septin 7a |
| chr8_+_21321765 | 0.06 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr22_-_36705684 | 0.06 |
|
|
|
| chr24_-_13118075 | 0.05 |
|
|
|
| chr21_+_10699071 | 0.05 |
|
|
|
| chr25_+_15901662 | 0.05 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr16_-_50139709 | 0.05 |
|
|
|
| chr20_-_36905932 | 0.04 |
ENSDART00000166966
|
slc25a27
|
solute carrier family 25, member 27 |
| chr23_-_17174766 | 0.04 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr16_-_22947908 | 0.04 |
ENSDART00000133910
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr1_+_28464109 | 0.04 |
ENSDART00000085648
|
lig4
|
ligase IV, DNA, ATP-dependent |
| chr21_-_25765319 | 0.04 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr5_-_37521979 | 0.03 |
ENSDART00000141791
ENSDART00000170528 |
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr5_+_60871396 | 0.03 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr4_+_16721069 | 0.03 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr7_-_25991514 | 0.03 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr23_+_36208651 | 0.02 |
ENSDART00000127174
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr4_+_23396557 | 0.02 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr23_+_22052045 | 0.02 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr10_-_35377693 | 0.02 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr2_-_22173254 | 0.01 |
ENSDART00000137538
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr17_+_15425559 | 0.01 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr7_-_29300402 | 0.01 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr25_-_19951796 | 0.01 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr2_+_22196842 | 0.01 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr19_-_7606443 | 0.00 |
ENSDART00000092256
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr13_+_18130077 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.5 | 3.8 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.3 | 1.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.6 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.1 | 0.8 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.7 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.5 | GO:0070572 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 1.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 1.8 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0010737 | protein kinase A signaling(GO:0010737) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 2.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 3.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |