Project
DANIO-CODE
Navigation
Downloads
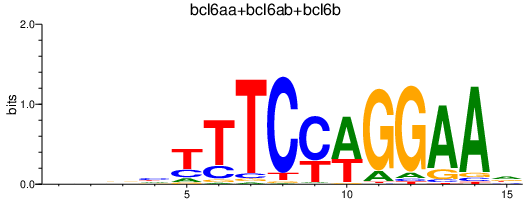
Results for bcl6aa+bcl6ab+bcl6b
Z-value: 1.26
Transcription factors associated with bcl6aa+bcl6ab+bcl6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl6ab
|
ENSDARG00000069295 | BCL6A transcription repressor b |
|
bcl6b
|
ENSDARG00000069335 | BCL6B transcription repressor |
|
bcl6aa
|
ENSDARG00000070864 | BCL6A transcription repressor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl6a | dr10_dc_chr6_-_28231995_28231999 | 0.27 | 3.1e-01 | Click! |
| bcl6ab | dr10_dc_chr2_-_10300329_10300384 | -0.14 | 6.1e-01 | Click! |
| bcl6b | dr10_dc_chr7_+_19765237_19765243 | -0.09 | 7.5e-01 | Click! |
Activity profile of bcl6aa+bcl6ab+bcl6b motif
Sorted Z-values of bcl6aa+bcl6ab+bcl6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl6aa+bcl6ab+bcl6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_17829889 | 3.26 |
ENSDART00000018948
ENSDART00000172343 |
cishb
|
cytokine inducible SH2-containing protein b |
| chr16_-_45950530 | 3.06 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr7_+_44373815 | 2.98 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr11_+_43083105 | 2.88 |
ENSDART00000130131
|
sult6b1
|
sulfotransferase family, cytosolic, 6b, member 1 |
| chr23_+_42431407 | 2.61 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr14_-_3264798 | 1.95 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr11_-_42934175 | 1.85 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr15_-_40391882 | 1.70 |
|
|
|
| chr24_-_2830194 | 1.58 |
ENSDART00000164913
|
si:ch211-152c8.5
|
si:ch211-152c8.5 |
| chr1_-_50147413 | 1.54 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr16_-_14973966 | 1.49 |
|
|
|
| chr2_+_314278 | 1.47 |
ENSDART00000155595
|
zgc:174263
|
zgc:174263 |
| chr11_-_22143133 | 1.43 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr17_-_30846176 | 1.41 |
ENSDART00000062811
|
yy1a
|
YY1 transcription factor a |
| chr4_-_72097569 | 1.33 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr21_-_2191847 | 1.32 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr11_+_2306187 | 1.29 |
|
|
|
| chr8_+_32604349 | 1.26 |
ENSDART00000126833
|
hmcn2
|
hemicentin 2 |
| chr21_+_35181329 | 1.24 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr3_-_60522527 | 1.22 |
ENSDART00000166334
|
srsf2b
|
serine/arginine-rich splicing factor 2b |
| chr18_+_22120282 | 1.21 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr7_-_22519196 | 1.17 |
ENSDART00000129919
|
ENSDARG00000091111
|
ENSDARG00000091111 |
| chr11_-_8158013 | 1.14 |
ENSDART00000024046
|
uox
|
urate oxidase |
| chr15_-_29454583 | 1.13 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr14_-_11765407 | 1.11 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr11_-_17829827 | 1.10 |
ENSDART00000018948
ENSDART00000172343 |
cishb
|
cytokine inducible SH2-containing protein b |
| chr19_+_30212276 | 1.07 |
|
|
|
| chr14_-_31128891 | 1.01 |
|
|
|
| chr1_+_27174549 | 1.01 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr18_-_36956268 | 1.00 |
|
|
|
| chr13_-_37482466 | 0.99 |
ENSDART00000147934
|
zgc:123068
|
zgc:123068 |
| chr3_+_39514598 | 0.97 |
ENSDART00000156038
|
epn2
|
epsin 2 |
| chr7_+_5408319 | 0.94 |
ENSDART00000142709
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr14_+_11151485 | 0.93 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr10_+_5268009 | 0.93 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr2_-_10919978 | 0.93 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr23_-_33631518 | 0.92 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr5_-_42272455 | 0.90 |
ENSDART00000161586
|
flot2a
|
flotillin 2a |
| chr21_-_2212429 | 0.88 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr13_+_46637827 | 0.88 |
ENSDART00000167931
|
mtrf1l
|
mitochondrial translational release factor 1-like |
| chr1_-_57931565 | 0.85 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr2_-_21960233 | 0.85 |
ENSDART00000027587
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr17_+_1080921 | 0.83 |
|
|
|
| chr14_+_6122948 | 0.83 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr20_+_10179249 | 0.82 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr7_+_32097831 | 0.81 |
ENSDART00000169588
|
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr23_-_19789409 | 0.81 |
ENSDART00000131860
|
rpl10
|
ribosomal protein L10 |
| chr23_+_42449089 | 0.79 |
ENSDART00000171119
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr1_-_25704525 | 0.79 |
ENSDART00000171614
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr11_-_42933969 | 0.79 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr11_+_43083214 | 0.77 |
ENSDART00000130131
|
sult6b1
|
sulfotransferase family, cytosolic, 6b, member 1 |
| chr25_+_35791595 | 0.76 |
ENSDART00000103013
|
ENSDARG00000070286
|
ENSDARG00000070286 |
| chr18_+_22120507 | 0.76 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr11_-_18649022 | 0.74 |
|
|
|
| chr1_+_43352429 | 0.70 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr24_-_37800277 | 0.69 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr1_-_20131393 | 0.69 |
|
|
|
| chr22_-_37676744 | 0.69 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr5_+_4033390 | 0.68 |
ENSDART00000149185
|
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr17_-_32471070 | 0.66 |
|
|
|
| chr25_+_35788501 | 0.64 |
ENSDART00000121838
|
hist1h4l
|
histone 1, H4, like |
| chr21_-_26078089 | 0.63 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr24_-_33377293 | 0.63 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr22_-_37676556 | 0.62 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr19_+_42936516 | 0.62 |
ENSDART00000165747
|
BX950855.1
|
ENSDARG00000103815 |
| chr7_-_6289729 | 0.61 |
ENSDART00000173323
|
hist1h4l
|
histone 1, H4, like |
| chr10_+_17470158 | 0.60 |
|
|
|
| chr8_-_23218886 | 0.60 |
ENSDART00000131632
|
ptk6a
|
PTK6 protein tyrosine kinase 6a |
| chr20_-_23949621 | 0.59 |
|
|
|
| chr14_+_11089382 | 0.57 |
ENSDART00000106657
|
glod5
|
glyoxalase domain containing 5 |
| chr1_-_54331157 | 0.55 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr25_-_34632441 | 0.55 |
ENSDART00000111724
|
hist1h4l
|
histone 1, H4, like |
| chr21_+_35181410 | 0.53 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr23_-_1024735 | 0.52 |
|
|
|
| chr18_-_46185361 | 0.49 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr23_+_42381296 | 0.49 |
ENSDART00000158684
ENSDART00000159985 ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr13_+_33578737 | 0.48 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr10_+_17470092 | 0.48 |
|
|
|
| chr19_+_30212487 | 0.48 |
|
|
|
| chr16_+_17808801 | 0.47 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr19_+_30212565 | 0.47 |
|
|
|
| chr10_+_5267746 | 0.46 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr7_-_22519593 | 0.46 |
ENSDART00000173496
|
ENSDARG00000091111
|
ENSDARG00000091111 |
| chr15_-_5811394 | 0.45 |
ENSDART00000158522
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr23_+_19743720 | 0.44 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr20_+_25669615 | 0.44 |
ENSDART00000063107
|
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr1_-_49028598 | 0.43 |
|
|
|
| chr5_-_64315770 | 0.41 |
ENSDART00000169287
|
zgc:110283
|
zgc:110283 |
| chr20_-_25726868 | 0.41 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr14_+_6122901 | 0.40 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr6_-_59510397 | 0.40 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr6_+_37677155 | 0.39 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr21_-_41191143 | 0.39 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr2_+_14016329 | 0.38 |
|
|
|
| chr18_+_18874077 | 0.37 |
ENSDART00000091094
|
pllp
|
plasmolipin |
| chr21_+_15660541 | 0.37 |
ENSDART00000101956
ENSDART00000026903 |
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr18_+_36085169 | 0.37 |
ENSDART00000059347
|
bckdha
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr21_+_15774822 | 0.37 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr4_-_18447913 | 0.36 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr8_-_47156727 | 0.36 |
|
|
|
| chr14_-_6680528 | 0.36 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr16_+_10438774 | 0.36 |
ENSDART00000121966
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr15_-_5811458 | 0.35 |
ENSDART00000158522
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr19_+_28363333 | 0.35 |
|
|
|
| chr19_+_5501487 | 0.34 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr10_+_5267937 | 0.34 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr24_+_42884 | 0.33 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr15_+_23615727 | 0.33 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr8_+_47108500 | 0.32 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr14_-_5371581 | 0.32 |
ENSDART00000012116
|
tlx2
|
T-cell leukemia, homeobox 2 |
| chr10_+_17470024 | 0.32 |
|
|
|
| chr8_+_47108614 | 0.32 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr11_+_30073930 | 0.31 |
ENSDART00000127075
|
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr13_-_49921086 | 0.31 |
ENSDART00000165066
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr16_+_46327528 | 0.31 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr21_+_15774697 | 0.30 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr8_-_28323246 | 0.30 |
ENSDART00000005583
|
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr20_+_32594366 | 0.30 |
ENSDART00000145175
|
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr15_+_20303330 | 0.30 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr15_+_20303673 | 0.30 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr19_+_26386711 | 0.29 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr5_+_23555868 | 0.29 |
ENSDART00000124379
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr5_+_36055362 | 0.29 |
ENSDART00000015240
|
capns1a
|
calpain, small subunit 1 a |
| chr24_-_18302095 | 0.28 |
ENSDART00000144203
ENSDART00000146024 |
sec61g
|
Sec61 translocon gamma subunit |
| chr14_+_7626822 | 0.28 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr22_+_26543146 | 0.28 |
|
|
|
| chr21_+_44586326 | 0.28 |
|
|
|
| chr2_-_10192027 | 0.28 |
ENSDART00000100709
|
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr14_-_1187074 | 0.27 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr10_-_42318897 | 0.27 |
ENSDART00000134890
ENSDART00000099259 |
dusp11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr20_+_25726978 | 0.27 |
ENSDART00000164264
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr16_+_17808913 | 0.27 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr6_-_51538315 | 0.27 |
|
|
|
| chr19_+_14490203 | 0.26 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr15_+_19389071 | 0.26 |
ENSDART00000164184
|
vps26b
|
VPS26 retromer complex component B |
| chr8_-_23762309 | 0.26 |
ENSDART00000141879
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr23_+_10211543 | 0.26 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr4_-_72115050 | 0.25 |
ENSDART00000174274
|
BX855614.4
|
ENSDARG00000105816 |
| chr6_-_28231995 | 0.25 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr25_-_16547149 | 0.25 |
ENSDART00000104110
|
slc25a18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr15_+_23861042 | 0.25 |
|
|
|
| chr5_+_36295353 | 0.25 |
ENSDART00000135776
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr6_+_26957509 | 0.25 |
ENSDART00000153595
|
farp2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr13_-_36996246 | 0.25 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr14_-_38469172 | 0.24 |
ENSDART00000159159
|
ENSDARG00000100564
|
ENSDARG00000100564 |
| chr11_-_2223434 | 0.24 |
|
|
|
| chr15_+_19388905 | 0.24 |
ENSDART00000149926
|
vps26b
|
VPS26 retromer complex component B |
| chr13_-_31165867 | 0.23 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr19_-_48144752 | 0.23 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr18_-_14891913 | 0.23 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr14_-_1186858 | 0.22 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr14_-_1186789 | 0.22 |
|
|
|
| chr16_+_16941228 | 0.22 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr1_+_39545535 | 0.22 |
ENSDART00000045697
|
zgc:56493
|
zgc:56493 |
| chr10_-_40904685 | 0.21 |
|
|
|
| chr6_+_7376428 | 0.21 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr4_+_72666635 | 0.21 |
ENSDART00000166994
|
trim24
|
tripartite motif containing 24 |
| chr7_-_981435 | 0.21 |
ENSDART00000101799
|
alpk1
|
alpha-kinase 1 |
| chr2_+_30911143 | 0.21 |
ENSDART00000132645
|
chmp5a
|
charged multivesicular body protein 5a |
| chr11_-_36088889 | 0.20 |
ENSDART00000146495
|
psma5
|
proteasome subunit alpha 5 |
| chr13_-_37482524 | 0.20 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr24_+_39139469 | 0.19 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr17_-_21258714 | 0.19 |
ENSDART00000157518
|
hspa12a
|
heat shock protein 12A |
| chr6_-_39524075 | 0.19 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr13_+_49527181 | 0.19 |
ENSDART00000176643
|
CABZ01084653.1
|
ENSDARG00000106801 |
| chr25_+_18487408 | 0.18 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr20_-_34865344 | 0.18 |
ENSDART00000142482
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr16_+_42868905 | 0.18 |
ENSDART00000156601
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr9_+_19655550 | 0.18 |
ENSDART00000111973
|
pde9a
|
phosphodiesterase 9A |
| chr18_-_44539130 | 0.18 |
|
|
|
| chr14_-_6669062 | 0.17 |
|
|
|
| chr19_+_38580804 | 0.17 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr16_-_1736470 | 0.17 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr17_+_16082472 | 0.16 |
ENSDART00000133154
|
znf395a
|
zinc finger protein 395a |
| chr7_+_30516734 | 0.16 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr6_-_34237257 | 0.16 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr25_+_4490129 | 0.16 |
|
|
|
| chr24_-_24023167 | 0.16 |
ENSDART00000080671
|
eif1axb
|
eukaryotic translation initiation factor 1A, X-linked, b |
| chr17_+_31205274 | 0.16 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr12_-_4584327 | 0.15 |
|
|
|
| chr11_-_1792616 | 0.15 |
|
|
|
| chr8_+_26037704 | 0.14 |
|
|
|
| chr21_+_3017623 | 0.14 |
|
|
|
| chr11_-_2274344 | 0.14 |
ENSDART00000158266
ENSDART00000104570 |
znf740a
|
zinc finger protein 740a |
| chr23_+_1024493 | 0.14 |
ENSDART00000143391
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr13_-_25418278 | 0.14 |
ENSDART00000077655
ENSDART00000168099 ENSDART00000135788 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr13_+_25418877 | 0.14 |
ENSDART00000169199
|
sec23ip
|
SEC23 interacting protein |
| chr6_-_40469263 | 0.13 |
ENSDART00000062724
|
crbn
|
cereblon |
| chr19_+_48144910 | 0.13 |
ENSDART00000149705
|
cdca8
|
cell division cycle associated 8 |
| chr8_-_47156995 | 0.13 |
|
|
|
| chr8_+_47909655 | 0.13 |
|
|
|
| chr1_-_58183299 | 0.12 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr19_+_38580844 | 0.12 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr14_-_6680812 | 0.12 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr5_-_32282404 | 0.12 |
ENSDART00000166698
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr23_+_2614658 | 0.12 |
|
|
|
| chr14_+_7626683 | 0.12 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr16_-_4780901 | 0.12 |
ENSDART00000029485
|
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr4_+_5634025 | 0.11 |
ENSDART00000028941
|
mrps18a
|
mitochondrial ribosomal protein S18A |
| chr17_+_10161999 | 0.11 |
ENSDART00000170274
|
foxa1
|
forkhead box A1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.3 | 0.9 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.3 | 3.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 0.9 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.9 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.2 | 0.7 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 4.0 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 1.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) abducens nucleus development(GO:0021742) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 3.0 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.3 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 1.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.6 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 2.7 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 1.7 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.2 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 2.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) muscle hypertrophy(GO:0014896) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 1.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) cGMP metabolic process(GO:0046068) |
| 0.0 | 0.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0006226 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.0 | 1.8 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.9 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.1 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0071256 | translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.7 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 1.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 1.3 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 3.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.1 | 0.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 3.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 2.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |