Project
DANIO-CODE
Navigation
Downloads
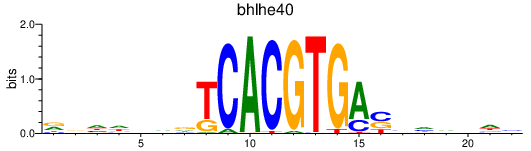
Results for bhlhe40
Z-value: 0.56
Transcription factors associated with bhlhe40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bhlhe40
|
ENSDARG00000004060 | basic helix-loop-helix family, member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bhlhe40 | dr10_dc_chr11_-_35501227_35501356 | -0.86 | 2.0e-05 | Click! |
Activity profile of bhlhe40 motif
Sorted Z-values of bhlhe40 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of bhlhe40
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_15536640 | 2.10 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr17_+_32670368 | 1.25 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| chr7_+_22027528 | 1.07 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr23_+_26153043 | 1.03 |
ENSDART00000054013
|
atp6ap1b
|
ATPase, H+ transporting, lysosomal accessory protein 1b |
| chr5_-_56539892 | 0.94 |
|
|
|
| chr12_+_18407380 | 0.94 |
ENSDART00000146260
|
msrb1b
|
methionine sulfoxide reductase B1b |
| chr9_-_34459799 | 0.94 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr3_+_16772351 | 0.92 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr9_-_10833577 | 0.87 |
ENSDART00000078348
ENSDART00000134911 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr3_+_13780104 | 0.86 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr24_+_12845339 | 0.85 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr10_+_32107014 | 0.82 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr12_+_18407435 | 0.78 |
ENSDART00000146260
|
msrb1b
|
methionine sulfoxide reductase B1b |
| chr23_+_35748728 | 0.74 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr25_-_19510499 | 0.72 |
ENSDART00000166824
|
gtse1
|
G-2 and S-phase expressed 1 |
| chr6_+_40924559 | 0.71 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_+_56539999 | 0.71 |
ENSDART00000167660
|
pja2
|
praja ring finger 2 |
| chr7_+_22027602 | 0.69 |
ENSDART00000056790
|
tmem256
|
transmembrane protein 256 |
| chr20_-_51287350 | 0.69 |
ENSDART00000023488
|
atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr19_+_791656 | 0.68 |
ENSDART00000138406
|
tmem79a
|
transmembrane protein 79a |
| chr10_-_44441390 | 0.68 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr1_-_46423056 | 0.65 |
ENSDART00000053157
|
setd4
|
SET domain containing 4 |
| chr14_-_36004908 | 0.65 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr9_-_34459768 | 0.64 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr8_-_47339228 | 0.64 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr15_+_1569674 | 0.63 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr5_+_25604404 | 0.63 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr16_+_25381315 | 0.62 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr10_-_44441197 | 0.62 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr6_-_49548896 | 0.61 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr12_+_47097965 | 0.60 |
ENSDART00000084714
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr4_+_17666992 | 0.60 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr12_-_7246994 | 0.58 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr8_-_47339136 | 0.56 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr11_+_5553243 | 0.55 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr9_-_10833656 | 0.55 |
ENSDART00000078348
ENSDART00000134911 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr16_+_8067699 | 0.54 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr6_+_131451 | 0.53 |
ENSDART00000166361
|
fdx1l
|
ferredoxin 1-like |
| chr18_+_14715573 | 0.52 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr3_+_13780013 | 0.51 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr6_-_49548929 | 0.48 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr17_+_30826890 | 0.46 |
ENSDART00000149600
|
tpp1
|
tripeptidyl peptidase I |
| chr3_+_34051311 | 0.45 |
ENSDART00000055252
|
TIMM29
|
translocase of inner mitochondrial membrane 29 |
| chr21_+_43183835 | 0.44 |
ENSDART00000175107
|
aff4
|
AF4/FMR2 family, member 4 |
| chr25_-_19510386 | 0.41 |
ENSDART00000147223
|
gtse1
|
G-2 and S-phase expressed 1 |
| chr25_-_35094522 | 0.40 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr13_+_9852854 | 0.40 |
ENSDART00000022051
|
gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr16_+_19223683 | 0.38 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr18_-_35152017 | 0.37 |
|
|
|
| chr3_-_25290808 | 0.37 |
ENSDART00000154200
|
bptf
|
bromodomain PHD finger transcription factor |
| chr17_-_15221715 | 0.36 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr19_+_40792676 | 0.36 |
ENSDART00000110699
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr1_-_46423011 | 0.34 |
ENSDART00000144899
|
setd4
|
SET domain containing 4 |
| chr7_-_41601001 | 0.34 |
ENSDART00000174258
|
zgc:92818
|
zgc:92818 |
| chr20_-_13244237 | 0.34 |
ENSDART00000124470
|
ints7
|
integrator complex subunit 7 |
| chr9_-_34459378 | 0.32 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr8_-_47339092 | 0.32 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr17_-_1949701 | 0.32 |
ENSDART00000013093
ENSDART00000168231 ENSDART00000166833 |
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr4_-_9195261 | 0.31 |
ENSDART00000080943
|
hcfc2
|
host cell factor C2 |
| chr23_-_31986679 | 0.31 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr15_-_41849769 | 0.30 |
ENSDART00000156819
|
slc25a36b
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36b |
| chr6_-_40924459 | 0.29 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr18_+_41521876 | 0.28 |
ENSDART00000136692
|
ccnl1a
|
cyclin L1a |
| chr13_+_40566900 | 0.28 |
ENSDART00000057047
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr5_+_59394192 | 0.27 |
|
|
|
| chr5_-_21627098 | 0.27 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr7_+_28782951 | 0.26 |
|
|
|
| chr23_-_10851353 | 0.26 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr13_+_29161591 | 0.26 |
ENSDART00000115023
|
PARG
|
poly(ADP-ribose) glycohydrolase |
| chr3_+_12633142 | 0.25 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr20_+_40869277 | 0.24 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr3_+_35310412 | 0.23 |
ENSDART00000176700
|
BX511029.1
|
ENSDARG00000107881 |
| chr25_+_3662390 | 0.23 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr20_+_14073428 | 0.22 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr17_+_30827047 | 0.22 |
ENSDART00000148547
|
tpp1
|
tripeptidyl peptidase I |
| chr15_+_1569864 | 0.18 |
ENSDART00000056765
|
smc4
|
structural maintenance of chromosomes 4 |
| chr5_+_59394222 | 0.16 |
|
|
|
| chr16_+_6789780 | 0.16 |
ENSDART00000149742
|
znf407
|
zinc finger protein 407 |
| chr16_+_8067533 | 0.15 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr21_+_43183570 | 0.15 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr19_+_9192592 | 0.13 |
ENSDART00000060442
|
setdb1a
|
SET domain, bifurcated 1a |
| chr19_+_7255491 | 0.13 |
ENSDART00000151220
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr20_-_51287382 | 0.13 |
ENSDART00000023488
|
atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr24_+_33548367 | 0.12 |
ENSDART00000166666
|
zgc:101800
|
zgc:101800 |
| chr20_+_14073464 | 0.12 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr20_-_29780783 | 0.11 |
ENSDART00000131219
ENSDART00000015928 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr20_-_40869235 | 0.10 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr5_+_19506512 | 0.10 |
ENSDART00000047841
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr3_+_16772396 | 0.09 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr8_-_25797849 | 0.08 |
ENSDART00000099364
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr11_+_5553275 | 0.08 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr15_-_44054313 | 0.07 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr15_-_26638024 | 0.07 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr21_+_11824091 | 0.06 |
ENSDART00000136103
|
ubap2a
|
ubiquitin associated protein 2a |
| chr17_-_37351769 | 0.06 |
ENSDART00000153522
|
ptgr2
|
prostaglandin reductase 2 |
| chr5_-_62762761 | 0.05 |
ENSDART00000163246
|
cntrl
|
centriolin |
| chr19_+_40792612 | 0.05 |
ENSDART00000017917
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr23_+_26152909 | 0.03 |
ENSDART00000129617
|
atp6ap1b
|
ATPase, H+ transporting, lysosomal accessory protein 1b |
| chr13_+_11741597 | 0.02 |
|
|
|
| chr9_+_16846294 | 0.01 |
ENSDART00000110866
|
cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr23_+_35748258 | 0.00 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 0.6 | GO:0071947 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.0 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.1 | 0.4 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 1.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.9 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.4 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 1.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.8 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 1.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.1 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.2 | GO:0005764 | lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.6 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |