Project
DANIO-CODE
Navigation
Downloads
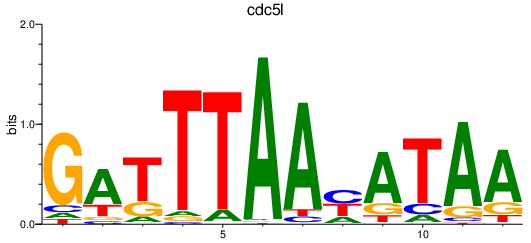
Results for cdc5l
Z-value: 1.14
Transcription factors associated with cdc5l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cdc5l
|
ENSDARG00000043797 | CDC5 cell division cycle 5-like (S. pombe) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cdc5l | dr10_dc_chr17_+_4902984_4903079 | 0.83 | 7.7e-05 | Click! |
Activity profile of cdc5l motif
Sorted Z-values of cdc5l motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cdc5l
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_35535503 | 3.13 |
ENSDART00000153249
|
tdrd6
|
tudor domain containing 6 |
| chr2_-_57339717 | 2.30 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr25_+_17764396 | 2.23 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr10_+_37457062 | 2.16 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr6_-_8008902 | 2.12 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr19_-_4206333 | 2.11 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr17_+_15527923 | 2.04 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr19_-_28004606 | 2.02 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr10_+_37457234 | 1.93 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr24_-_14446593 | 1.83 |
|
|
|
| chr9_+_34588426 | 1.82 |
ENSDART00000011304
|
med14
|
mediator complex subunit 14 |
| chr23_+_24672435 | 1.75 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr25_+_17829003 | 1.72 |
ENSDART00000067305
|
zgc:103499
|
zgc:103499 |
| chr6_-_42371527 | 1.66 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr18_+_42925672 | 1.64 |
|
|
|
| chr7_+_13329491 | 1.63 |
ENSDART00000158797
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr6_+_40995061 | 1.62 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr11_-_18112090 | 1.61 |
ENSDART00000019248
|
tmem110
|
transmembrane protein 110 |
| chr4_+_16020464 | 1.60 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr16_+_29574449 | 1.59 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr1_+_11508857 | 1.59 |
ENSDART00000172334
ENSDART00000112171 |
tdrd7a
|
tudor domain containing 7 a |
| chr12_-_17741377 | 1.58 |
ENSDART00000166604
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr25_-_36621626 | 1.57 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr23_+_43868027 | 1.56 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr5_+_65453249 | 1.55 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr24_-_10253851 | 1.54 |
ENSDART00000127568
ENSDART00000106260 |
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr24_+_39237314 | 1.51 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr19_+_2942485 | 1.50 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr6_+_40994859 | 1.50 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr22_-_10861268 | 1.47 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr3_-_54352535 | 1.44 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr16_-_21734502 | 1.41 |
ENSDART00000179125
|
CABZ01009710.1
|
ENSDARG00000106589 |
| chr5_-_14993912 | 1.41 |
ENSDART00000085943
|
taok3a
|
TAO kinase 3a |
| chr18_+_6396038 | 1.40 |
ENSDART00000024576
|
fkbp4
|
FK506 binding protein 4 |
| chr3_+_17562348 | 1.38 |
|
|
|
| chr10_-_22180658 | 1.37 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr6_+_40994918 | 1.37 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr9_-_21287478 | 1.36 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr5_-_61654526 | 1.33 |
ENSDART00000158570
|
zgc:85789
|
zgc:85789 |
| chr1_+_44516337 | 1.33 |
ENSDART00000125037
|
EVI5L
|
ecotropic viral integration site 5 like |
| chr21_-_31189338 | 1.33 |
ENSDART00000011572
|
zgc:152891
|
zgc:152891 |
| chr22_-_21021942 | 1.30 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr9_+_45193290 | 1.30 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr17_+_31722625 | 1.30 |
ENSDART00000156180
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr24_+_31522539 | 1.29 |
ENSDART00000126380
|
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr15_-_14102102 | 1.29 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr4_-_13025193 | 1.28 |
ENSDART00000177894
|
BX119963.3
|
ENSDARG00000106390 |
| chr6_-_8009143 | 1.28 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr19_+_7082199 | 1.25 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr4_+_9466175 | 1.25 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr14_-_6901209 | 1.25 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_-_26654610 | 1.24 |
ENSDART00000134448
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr23_+_27777325 | 1.24 |
ENSDART00000134008
|
lmbr1l
|
limb development membrane protein 1-like |
| chr19_+_3115685 | 1.24 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr17_+_23534953 | 1.24 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr2_-_19585211 | 1.23 |
ENSDART00000169384
|
cdc20
|
cell division cycle 20 homolog |
| chr10_+_10354740 | 1.22 |
ENSDART00000142895
|
urm1
|
ubiquitin related modifier 1 |
| chr8_-_3354396 | 1.22 |
ENSDART00000169430
ENSDART00000167187 ENSDART00000170478 |
FUT9 (1 of many)
fut9b
|
fucosyltransferase 9 fucosyltransferase 9b |
| chr8_-_16689736 | 1.21 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr18_+_6396115 | 1.21 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr1_+_21244200 | 1.20 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr19_+_30862829 | 1.19 |
ENSDART00000073704
|
si:ch211-215a10.4
|
si:ch211-215a10.4 |
| chr17_-_43542320 | 1.19 |
ENSDART00000154595
|
si:dkey-21a6.5
|
si:dkey-21a6.5 |
| chr5_-_8712114 | 1.17 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr7_-_20212157 | 1.17 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr10_-_33307992 | 1.17 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr9_+_34588788 | 1.16 |
ENSDART00000146314
|
med14
|
mediator complex subunit 14 |
| chr8_-_3974320 | 1.16 |
ENSDART00000159895
|
mtmr3
|
myotubularin related protein 3 |
| chr7_-_31346802 | 1.16 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr16_-_39291647 | 1.14 |
ENSDART00000171342
|
tmem42a
|
transmembrane protein 42a |
| chr25_-_15176295 | 1.14 |
|
|
|
| chr2_-_22875246 | 1.14 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr9_+_30250973 | 1.13 |
|
|
|
| chr16_-_52847564 | 1.13 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr19_+_16159980 | 1.12 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr25_-_3091345 | 1.10 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr7_-_20212038 | 1.09 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr14_+_31265616 | 1.09 |
|
|
|
| chr7_+_38417230 | 1.09 |
ENSDART00000162479
|
BX957297.1
|
ENSDARG00000098951 |
| chr13_-_6150836 | 1.06 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr8_-_53165501 | 1.06 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr3_-_54591713 | 1.05 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr19_+_3115791 | 1.05 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr19_+_7489293 | 1.05 |
ENSDART00000081746
|
apoa1bp
|
apolipoprotein A-I binding protein |
| chr3_-_18039574 | 1.03 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr2_+_30198819 | 1.02 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr8_+_10825036 | 1.02 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr23_+_24672673 | 1.02 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr1_+_40134345 | 1.01 |
ENSDART00000177517
|
CR387992.1
|
ENSDARG00000108790 |
| chr2_+_5383347 | 1.01 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr6_-_8468961 | 1.00 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr12_+_6032235 | 1.00 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr5_-_8712068 | 1.00 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr18_+_18077901 | 1.00 |
|
|
|
| chr20_+_42640466 | 0.99 |
ENSDART00000134066
|
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr1_-_1204375 | 0.98 |
ENSDART00000165762
|
CR450787.1
|
ENSDARG00000103469 |
| chr5_-_9288622 | 0.98 |
ENSDART00000158874
|
ddr2l
|
discoidin domain receptor family, member 2, like |
| chr6_+_60130111 | 0.97 |
ENSDART00000148557
|
aurka
|
aurora kinase A |
| chr21_+_10692781 | 0.96 |
|
|
|
| chr9_+_24255311 | 0.96 |
ENSDART00000166303
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr11_-_30261689 | 0.96 |
ENSDART00000101667
|
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr3_-_20989306 | 0.96 |
|
|
|
| chr3_+_6557967 | 0.96 |
ENSDART00000162255
|
nup85
|
nucleoporin 85 |
| chr7_-_28376856 | 0.96 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr16_-_32254735 | 0.95 |
ENSDART00000023995
|
fam26e.2
|
family with sequence similarity 26, member E, tandem duplicate 2 |
| chr3_-_43253952 | 0.95 |
ENSDART00000167159
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr22_+_1905394 | 0.95 |
ENSDART00000165758
|
znf1181
|
zinc finger protein 1181 |
| chr16_-_43061368 | 0.95 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr14_+_21835107 | 0.94 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr25_-_28630138 | 0.94 |
|
|
|
| chr7_-_31346844 | 0.93 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr3_+_26113651 | 0.93 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr11_-_33605979 | 0.92 |
ENSDART00000171439
|
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr24_-_31967674 | 0.92 |
ENSDART00000156060
|
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr11_-_6870810 | 0.92 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr15_-_41732606 | 0.92 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr6_-_30696540 | 0.91 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr6_-_9394432 | 0.91 |
ENSDART00000013066
|
ercc3
|
excision repair cross-complementation group 3 |
| chr19_-_3115104 | 0.91 |
ENSDART00000093146
|
bop1
|
block of proliferation 1 |
| chr23_+_2104589 | 0.91 |
|
|
|
| chr2_+_30756519 | 0.91 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr7_-_61373495 | 0.90 |
ENSDART00000098622
|
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr5_-_18502344 | 0.90 |
ENSDART00000137022
ENSDART00000090494 ENSDART00000165701 |
golga3
|
golgin A3 |
| chr2_-_23735018 | 0.90 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr23_-_24616306 | 0.90 |
ENSDART00000088777
|
atp13a2
|
ATPase type 13A2 |
| chr22_+_1462211 | 0.89 |
|
|
|
| chr19_+_3115642 | 0.89 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr4_+_17653548 | 0.89 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr6_+_60130210 | 0.89 |
ENSDART00000148557
|
aurka
|
aurora kinase A |
| chr24_-_28358514 | 0.88 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr18_+_18890626 | 0.88 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr11_+_16017857 | 0.88 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr17_-_31061338 | 0.88 |
|
|
|
| chr20_+_43794079 | 0.88 |
ENSDART00000045185
|
lin9
|
lin-9 DREAM MuvB core complex component |
| chr2_+_52456425 | 0.87 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr12_+_35101949 | 0.87 |
ENSDART00000153022
|
AL935194.1
|
ENSDARG00000060678 |
| chr17_+_15528557 | 0.87 |
|
|
|
| chr16_+_19831573 | 0.86 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr24_+_37450437 | 0.86 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr1_+_23778514 | 0.85 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr24_-_23793117 | 0.85 |
|
|
|
| chr23_-_3778250 | 0.85 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr16_+_25248338 | 0.84 |
ENSDART00000105514
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr24_+_12845339 | 0.84 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr7_+_24610446 | 0.83 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr7_-_28376722 | 0.83 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr2_-_52156537 | 0.83 |
ENSDART00000168875
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr24_-_23639458 | 0.82 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr17_+_13078286 | 0.82 |
ENSDART00000149779
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr19_+_2942373 | 0.82 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr3_-_30955799 | 0.82 |
ENSDART00000103421
|
zgc:153292
|
zgc:153292 |
| chr4_+_25192397 | 0.82 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr2_+_30756137 | 0.82 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr10_-_1185831 | 0.81 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr11_+_1506660 | 0.81 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr11_+_1506720 | 0.80 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr1_+_21244242 | 0.80 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr24_-_28358595 | 0.80 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr15_-_45387456 | 0.80 |
ENSDART00000176116
|
CABZ01067262.1
|
ENSDARG00000107582 |
| chr2_+_23004159 | 0.80 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr4_+_17653739 | 0.79 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr16_-_46558057 | 0.79 |
ENSDART00000169477
|
tmem176l.1
|
transmembrane protein 176l.1 |
| chr7_+_39392939 | 0.79 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr10_+_21477579 | 0.79 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr4_-_75683584 | 0.78 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr23_-_24615681 | 0.78 |
ENSDART00000132265
|
atp13a2
|
ATPase type 13A2 |
| chr9_-_34450885 | 0.78 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr10_+_35209240 | 0.77 |
ENSDART00000126105
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr20_-_27291231 | 0.77 |
ENSDART00000020710
ENSDART00000149024 |
btbd7
|
BTB (POZ) domain containing 7 |
| chr20_+_22900165 | 0.77 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr14_-_20843701 | 0.77 |
ENSDART00000075759
|
maea
|
macrophage erythroblast attacher |
| chr7_+_10319999 | 0.76 |
ENSDART00000168801
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr16_-_43107682 | 0.76 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr6_+_40994992 | 0.75 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr10_-_33307845 | 0.75 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr18_+_27095471 | 0.75 |
ENSDART00000125326
ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr24_+_21030141 | 0.75 |
ENSDART00000154940
|
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_-_29958364 | 0.75 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr23_+_26099718 | 0.74 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_-_34293596 | 0.74 |
ENSDART00000128869
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr1_+_51216305 | 0.74 |
|
|
|
| chr21_-_22321244 | 0.74 |
ENSDART00000136904
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr11_-_38856849 | 0.74 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr18_-_18886144 | 0.74 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr22_-_10861329 | 0.73 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr9_+_23813071 | 0.73 |
|
|
|
| chr3_+_41984196 | 0.73 |
ENSDART00000154052
|
ftsj2
|
FtsJ RNA methyltransferase homolog 2 (E. coli) |
| chr24_-_16835352 | 0.73 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr5_-_23291839 | 0.73 |
ENSDART00000099083
ENSDART00000099084 |
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr19_+_7717962 | 0.73 |
ENSDART00000112404
|
cgnb
|
cingulin b |
| chr22_+_1894340 | 0.72 |
ENSDART00000164158
|
znf1156
|
zinc finger protein 1156 |
| chr21_-_22085596 | 0.72 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr16_-_32708605 | 0.72 |
ENSDART00000133351
|
coq3
|
coenzyme Q3 methyltransferase |
| chr4_-_13615927 | 0.72 |
ENSDART00000138366
|
irf5
|
interferon regulatory factor 5 |
| chr8_-_28255076 | 0.72 |
ENSDART00000050671
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr14_+_47190202 | 0.71 |
ENSDART00000074116
|
rab33ba
|
RAB33B, member RAS oncogene family a |
| chr18_+_27769446 | 0.71 |
|
|
|
| chr11_+_42433566 | 0.70 |
ENSDART00000162327
|
zgc:194981
|
zgc:194981 |
| chr22_+_14092247 | 0.70 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr18_-_5167807 | 0.70 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:2001284 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.7 | 2.7 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.7 | 5.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.5 | 1.5 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.4 | 4.8 | GO:0030719 | P granule organization(GO:0030719) |
| 0.4 | 1.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.4 | 1.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 4.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.3 | 1.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.3 | 1.3 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 1.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 1.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.3 | 1.4 | GO:0032516 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.3 | 0.8 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 1.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.2 | 0.7 | GO:0042128 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 0.6 | GO:1900543 | negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of purine nucleotide metabolic process(GO:1900543) |
| 0.2 | 2.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.6 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 1.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 1.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 0.7 | GO:0051591 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.2 | GO:1901906 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 1.2 | GO:0070601 | establishment of mitotic sister chromatid cohesion(GO:0034087) centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.3 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.1 | 1.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.6 | GO:1902592 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 3.6 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 0.7 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.5 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 3.7 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 3.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.7 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.6 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 2.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.7 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 1.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.6 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.1 | 0.6 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 1.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.7 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:1903335 | regulation of vacuolar transport(GO:1903335) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.2 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.1 | 0.2 | GO:0070973 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.6 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.6 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.5 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 1.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.9 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 2.0 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.7 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.3 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.0 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 1.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 1.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.7 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.8 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.7 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 1.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.7 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 1.2 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 2.6 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.3 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.6 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 2.8 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 1.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 3.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.4 | GO:0005675 | holo TFIIH complex(GO:0005675) transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 0.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 1.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.2 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 2.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 2.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.6 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.5 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.6 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 1.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 6.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.0 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 1.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.5 | 1.6 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.5 | 1.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.5 | 1.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.5 | 1.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.4 | 1.3 | GO:0030580 | 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity(GO:0008425) quinone cofactor methyltransferase activity(GO:0030580) |
| 0.4 | 1.6 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.4 | 5.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 2.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 0.8 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 0.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 1.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 4.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 0.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.7 | GO:0008942 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.2 | 1.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 2.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.2 | 2.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 1.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 1.0 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.2 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.1 | 0.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.6 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.3 | GO:0003823 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.1 | 0.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 3.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.8 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 3.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 3.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |