Project
DANIO-CODE
Navigation
Downloads
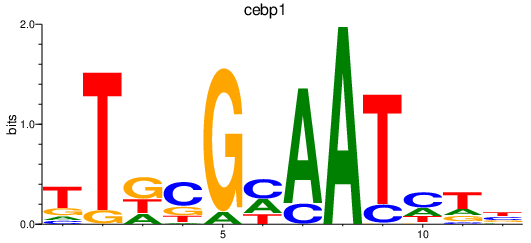
Results for cebp1
Z-value: 1.01
Transcription factors associated with cebp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebp1
|
ENSDARG00000101096 | CCAAT/enhancer binding protein (C/EBP) 1 |
Activity profile of cebp1 motif
Sorted Z-values of cebp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cebp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_17562348 | 2.85 |
|
|
|
| chr2_+_28199458 | 2.61 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr19_-_44459352 | 2.55 |
ENSDART00000151084
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr12_-_22116694 | 2.51 |
ENSDART00000038310
ENSDART00000132731 |
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr18_-_38289370 | 2.31 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr12_+_30673985 | 2.15 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr21_-_11562388 | 2.09 |
ENSDART00000139289
|
cast
|
calpastatin |
| chr16_-_46558057 | 1.90 |
ENSDART00000169477
|
tmem176l.1
|
transmembrane protein 176l.1 |
| chr14_+_34150130 | 1.84 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr10_+_37457234 | 1.77 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr7_+_58910115 | 1.76 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr24_-_7534334 | 1.74 |
|
|
|
| chr17_+_24791024 | 1.69 |
ENSDART00000130871
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr5_+_52199662 | 1.69 |
|
|
|
| chr1_-_23767216 | 1.68 |
ENSDART00000140197
|
sh3d19
|
SH3 domain containing 19 |
| chr14_+_35124708 | 1.68 |
ENSDART00000115307
|
ENSDARG00000079591
|
ENSDARG00000079591 |
| chr9_+_29737843 | 1.63 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr5_-_57053687 | 1.47 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr21_+_43183522 | 1.47 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr17_+_24790812 | 1.46 |
ENSDART00000082251
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr22_+_16070849 | 1.45 |
ENSDART00000165216
|
dph5
|
diphthamide biosynthesis 5 |
| chr10_+_6925373 | 1.44 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr19_-_35408237 | 1.43 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr6_+_44199738 | 1.43 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr6_+_422911 | 1.40 |
|
|
|
| chr7_+_37818298 | 1.39 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr17_-_11904068 | 1.38 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr9_+_18821640 | 1.37 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr14_+_35124867 | 1.29 |
ENSDART00000115307
|
ENSDARG00000079591
|
ENSDARG00000079591 |
| chr5_-_47323502 | 1.29 |
ENSDART00000175026
|
ccnh
|
cyclin H |
| chr8_+_9660595 | 1.28 |
ENSDART00000091848
|
gripap1
|
GRIP1 associated protein 1 |
| chr13_-_49934440 | 1.28 |
ENSDART00000034541
|
gpatch11
|
G patch domain containing 11 |
| chr14_+_35124913 | 1.24 |
ENSDART00000115307
|
ENSDARG00000079591
|
ENSDARG00000079591 |
| chr14_+_35084220 | 1.24 |
ENSDART00000171809
|
sytl4
|
synaptotagmin-like 4 |
| chr21_-_34227158 | 1.21 |
ENSDART00000169218
ENSDART00000064320 ENSDART00000172381 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr7_+_37818365 | 1.20 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr20_+_34091702 | 1.18 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr4_-_2319514 | 1.14 |
ENSDART00000150039
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr22_+_2049719 | 1.13 |
|
|
|
| chr6_+_23709605 | 1.11 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr24_+_26223767 | 1.11 |
|
|
|
| chr23_-_32267833 | 1.10 |
|
|
|
| chr13_-_3805904 | 1.10 |
ENSDART00000017052
|
ncoa4
|
nuclear receptor coactivator 4 |
| chr22_-_16351654 | 1.08 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr5_+_26888419 | 1.07 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr11_-_17668381 | 1.07 |
ENSDART00000080752
|
uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr21_-_11562321 | 1.05 |
ENSDART00000144370
|
cast
|
calpastatin |
| chr14_+_35124973 | 1.05 |
ENSDART00000115307
|
ENSDARG00000079591
|
ENSDARG00000079591 |
| chr10_+_21477579 | 1.04 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr6_+_40924559 | 1.03 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr17_+_32670368 | 1.03 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| chr9_+_30815655 | 1.02 |
ENSDART00000147813
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr24_-_32668102 | 1.02 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr5_-_14783255 | 1.02 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr6_+_54488452 | 1.00 |
ENSDART00000156263
|
ENSDARG00000044061
|
ENSDARG00000044061 |
| chr20_-_38884093 | 1.00 |
ENSDART00000153430
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr20_-_36776382 | 0.98 |
ENSDART00000062908
|
rpl7l1
|
ribosomal protein L7-like 1 |
| KN150335v1_-_42498 | 0.98 |
|
|
|
| chr8_+_20108592 | 0.97 |
|
|
|
| chr12_+_31668126 | 0.96 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr7_+_10320931 | 0.96 |
ENSDART00000173125
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr18_+_47305642 | 0.96 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr25_+_4888647 | 0.96 |
ENSDART00000165933
ENSDART00000159537 |
hdac10
|
histone deacetylase 10 |
| chr12_+_4885307 | 0.95 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr2_+_34675356 | 0.95 |
|
|
|
| chr12_+_31668168 | 0.95 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr25_+_18855868 | 0.95 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr9_+_34423285 | 0.94 |
ENSDART00000174944
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr10_-_44634167 | 0.94 |
ENSDART00000010056
ENSDART00000169919 |
ccdc62
|
coiled-coil domain containing 62 |
| chr9_-_32532843 | 0.93 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr17_-_31195376 | 0.93 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr20_-_31524684 | 0.93 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr15_+_6462943 | 0.93 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr12_+_4885359 | 0.93 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr25_+_5995708 | 0.93 |
ENSDART00000067514
|
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr6_+_46307828 | 0.92 |
ENSDART00000154817
|
si:dkeyp-67f1.1
|
si:dkeyp-67f1.1 |
| chr20_+_41351373 | 0.92 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| chr23_+_33792145 | 0.92 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr20_-_34126039 | 0.92 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr13_+_24266306 | 0.91 |
ENSDART00000101200
ENSDART00000166005 |
zgc:153169
|
zgc:153169 |
| chr9_-_32532691 | 0.90 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr2_+_223139 | 0.90 |
ENSDART00000113021
|
dhx30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr22_-_4605581 | 0.89 |
ENSDART00000028634
|
cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr16_-_12582192 | 0.88 |
|
|
|
| chr12_+_4885394 | 0.88 |
ENSDART00000159367
|
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr8_-_14013213 | 0.87 |
ENSDART00000133922
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr10_+_32122607 | 0.87 |
ENSDART00000062311
ENSDART00000124166 |
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr10_-_36849734 | 0.86 |
|
|
|
| chr9_+_19812730 | 0.86 |
ENSDART00000110009
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr5_+_27793171 | 0.85 |
|
|
|
| chr13_+_29904945 | 0.85 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr15_-_41732430 | 0.84 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr12_-_10327938 | 0.84 |
ENSDART00000007335
|
ndc80
|
NDC80 kinetochore complex component |
| chr11_-_33956671 | 0.84 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr15_+_35089305 | 0.83 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr20_-_34894607 | 0.83 |
|
|
|
| chr5_-_19459809 | 0.83 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr11_+_19440815 | 0.83 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr16_-_9939549 | 0.82 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr15_-_20476476 | 0.81 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr10_-_42279805 | 0.81 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr2_-_48345919 | 0.80 |
|
|
|
| chr24_+_37810878 | 0.80 |
ENSDART00000066556
|
zmp:0000000624
|
zmp:0000000624 |
| chr23_+_17596504 | 0.80 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr10_-_32716718 | 0.80 |
ENSDART00000063544
|
atg101
|
autophagy related 101 |
| chr11_+_24820942 | 0.79 |
|
|
|
| chr19_-_22762165 | 0.79 |
ENSDART00000169065
ENSDART00000080260 |
zgc:109744
|
zgc:109744 |
| chr12_-_21562929 | 0.79 |
ENSDART00000152999
|
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr23_-_29952020 | 0.79 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr5_-_26195227 | 0.79 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr19_+_29871997 | 0.78 |
|
|
|
| chr10_+_36752015 | 0.78 |
ENSDART00000171392
|
rab6a
|
RAB6A, member RAS oncogene family |
| chr19_-_35392661 | 0.78 |
ENSDART00000103665
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr11_+_12662209 | 0.77 |
ENSDART00000054837
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_-_28257339 | 0.77 |
ENSDART00000148331
|
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr5_-_19459698 | 0.76 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr1_+_30840599 | 0.76 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr18_-_35485549 | 0.76 |
ENSDART00000141023
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr5_-_11309164 | 0.76 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr25_+_11212787 | 0.75 |
ENSDART00000159583
|
FQ312013.1
|
ENSDARG00000099473 |
| chr22_-_18466730 | 0.75 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr12_-_13690539 | 0.74 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr9_+_2523927 | 0.74 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr10_-_36849596 | 0.74 |
|
|
|
| chr20_+_34598242 | 0.74 |
ENSDART00000143765
|
gorab
|
golgin, rab6-interacting |
| chr6_+_3313468 | 0.74 |
ENSDART00000011785
|
rad54l
|
RAD54-like (S. cerevisiae) |
| chr3_+_3826209 | 0.73 |
ENSDART00000150931
|
cdc42ep1b
|
CDC42 effector protein (Rho GTPase binding) 1b |
| chr8_-_16479425 | 0.73 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr19_-_431762 | 0.72 |
ENSDART00000165371
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr20_+_37023072 | 0.72 |
ENSDART00000155058
|
CR388421.1
|
ENSDARG00000096706 |
| chr20_-_20921480 | 0.72 |
ENSDART00000152781
|
ckbb
|
creatine kinase, brain b |
| chr13_+_29904913 | 0.71 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr4_+_18525469 | 0.71 |
ENSDART00000154154
|
BX649398.1
|
ENSDARG00000097195 |
| chr2_-_15380727 | 0.70 |
|
|
|
| chr15_-_41732606 | 0.70 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr23_-_32309547 | 0.70 |
ENSDART00000103343
|
grasp
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr5_-_35997345 | 0.70 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr15_-_41732304 | 0.70 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr17_+_51655501 | 0.70 |
ENSDART00000149807
|
odc1
|
ornithine decarboxylase 1 |
| chr10_-_42279667 | 0.70 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr19_+_31280451 | 0.69 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr19_-_22761674 | 0.67 |
|
|
|
| chr21_-_21491268 | 0.67 |
ENSDART00000014254
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr25_+_2252667 | 0.66 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr17_+_18012036 | 0.65 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr3_-_26060047 | 0.65 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr13_+_11307037 | 0.64 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr12_+_26971687 | 0.64 |
|
|
|
| chr17_-_31195299 | 0.64 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr13_+_7243353 | 0.64 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr20_-_38030337 | 0.63 |
ENSDART00000153451
|
angel2
|
angel homolog 2 (Drosophila) |
| chr3_-_26060098 | 0.63 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr21_-_26369503 | 0.63 |
ENSDART00000137312
|
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr11_+_12662507 | 0.63 |
ENSDART00000135761
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr20_+_29306863 | 0.62 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr25_-_36678422 | 0.62 |
ENSDART00000087247
|
glg1a
|
golgi glycoprotein 1a |
| chr8_-_11286437 | 0.61 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr22_+_7983496 | 0.61 |
|
|
|
| chr17_-_23874858 | 0.61 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr13_-_32324315 | 0.61 |
|
|
|
| chr19_+_32178335 | 0.61 |
|
|
|
| chr10_+_32122560 | 0.61 |
ENSDART00000062311
ENSDART00000124166 |
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr10_-_21971084 | 0.61 |
ENSDART00000174954
|
CABZ01031892.1
|
ENSDARG00000107280 |
| chr5_+_36067010 | 0.61 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr21_-_17388020 | 0.60 |
ENSDART00000101295
|
gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr13_+_3807780 | 0.60 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr8_+_31247312 | 0.59 |
ENSDART00000174921
|
AL929453.2
|
ENSDARG00000108092 |
| chr17_-_28794619 | 0.59 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr13_+_15685745 | 0.58 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr6_+_10098305 | 0.58 |
ENSDART00000151477
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr8_-_25696708 | 0.57 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr16_-_33864015 | 0.57 |
|
|
|
| chr15_-_21733882 | 0.57 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr12_-_45496246 | 0.57 |
ENSDART00000176229
ENSDART00000175457 |
FO818685.1
|
ENSDARG00000107895 |
| chr5_-_11309124 | 0.57 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr12_-_13690748 | 0.57 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr14_-_6220150 | 0.57 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr12_-_4597080 | 0.56 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr25_+_27301351 | 0.56 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr7_-_71338273 | 0.56 |
|
|
|
| chr20_+_41351477 | 0.56 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| chr6_-_10492724 | 0.56 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr23_-_43531724 | 0.56 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr8_-_11286308 | 0.55 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr5_-_29782972 | 0.55 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr7_-_18816332 | 0.55 |
ENSDART00000108787
|
ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr3_+_21905456 | 0.54 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr20_-_21906340 | 0.54 |
ENSDART00000145807
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr1_+_8010026 | 0.54 |
ENSDART00000152142
|
zgc:77849
|
zgc:77849 |
| chr19_-_30300240 | 0.53 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr14_-_26093882 | 0.53 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr21_+_17014240 | 0.53 |
ENSDART00000047201
|
atp2a2b
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2b |
| chr19_-_8849482 | 0.52 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr20_-_33584753 | 0.52 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr9_+_25109716 | 0.52 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_-_27395825 | 0.52 |
ENSDART00000170779
|
snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr25_+_5995507 | 0.52 |
ENSDART00000067514
|
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr5_+_13370060 | 0.52 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr10_+_44634362 | 0.52 |
|
|
|
| chr9_+_24255311 | 0.51 |
ENSDART00000166303
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.8 | 2.5 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.6 | 1.8 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.5 | 2.1 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 2.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 1.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.4 | 1.4 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.4 | 1.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 1.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 2.1 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.3 | 1.3 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.3 | 0.8 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 0.9 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 1.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 2.0 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 0.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 0.8 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 1.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 1.1 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.2 | 0.7 | GO:1990379 | lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.2 | 0.8 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.2 | 1.0 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 1.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.4 | GO:0008406 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.1 | 0.6 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 1.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 0.4 | GO:0045041 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 1.9 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.3 | GO:0050854 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.3 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.1 | 1.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.4 | GO:1902751 | regulation of mitotic cell cycle, embryonic(GO:0009794) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic cell cycle, embryonic(GO:0045448) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.9 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.2 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.8 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 1.6 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.6 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.1 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 0.3 | GO:1990481 | snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:0010984 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.7 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.7 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.7 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 1.1 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 4.3 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.7 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.8 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 1.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.0 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.6 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.9 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.9 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 1.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 1.0 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 1.3 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.7 | 2.6 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.5 | 2.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 1.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 1.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.3 | 1.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.3 | 0.8 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.3 | 0.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 0.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 1.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 0.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 2.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 0.5 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.7 | GO:0045495 | pole plasm(GO:0045495) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.3 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 5.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.4 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.4 | 1.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.3 | 1.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.3 | 1.0 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 1.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.2 | 2.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 0.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.9 | GO:0003948 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity(GO:0003948) |
| 0.2 | 0.7 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.2 | 1.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.8 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.3 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.1 | 0.5 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 1.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 1.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.2 | GO:0016844 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.1 | 2.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.2 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.8 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.4 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |