Project
DANIO-CODE
Navigation
Downloads
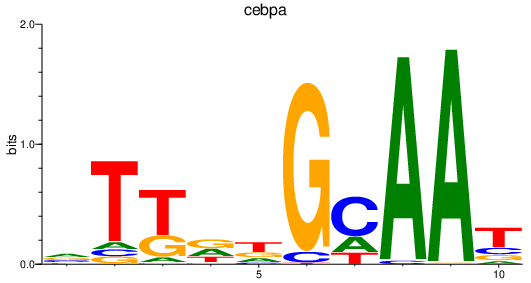
Results for cebpa
Z-value: 1.17
Transcription factors associated with cebpa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpa
|
ENSDARG00000036074 | CCAAT enhancer binding protein alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpa | dr10_dc_chr7_-_37816600_37816633 | 0.51 | 4.5e-02 | Click! |
Activity profile of cebpa motif
Sorted Z-values of cebpa motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpa
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_14970278 | 2.83 |
ENSDART00000003847
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr13_-_18506153 | 2.44 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr5_+_8415025 | 2.36 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr14_+_36545240 | 2.32 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr8_-_50159285 | 2.31 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr5_-_30015572 | 2.23 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr16_-_31670211 | 2.22 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr13_+_24148599 | 2.15 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr6_+_56157608 | 2.11 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_-_41194452 | 1.98 |
ENSDART00000165949
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr19_-_5781358 | 1.96 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr10_+_26020615 | 1.96 |
ENSDART00000128292
ENSDART00000170275 ENSDART00000166164 ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr16_-_21981065 | 1.93 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr25_+_30746445 | 1.93 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr7_+_6814828 | 1.82 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr15_+_32853646 | 1.82 |
ENSDART00000167515
|
postnb
|
periostin, osteoblast specific factor b |
| chr22_-_17570306 | 1.76 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr18_-_782643 | 1.75 |
ENSDART00000161084
|
si:dkey-205h23.1
|
si:dkey-205h23.1 |
| chr9_-_4890648 | 1.75 |
ENSDART00000167998
|
fmnl2a
|
formin-like 2a |
| chr1_+_9367672 | 1.74 |
ENSDART00000144756
|
fgb
|
fibrinogen beta chain |
| chr2_+_24852094 | 1.71 |
ENSDART00000052063
|
rps28
|
ribosomal protein S28 |
| chr25_+_4918339 | 1.68 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr25_+_7660590 | 1.66 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr20_-_52459338 | 1.62 |
ENSDART00000135463
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr5_+_26195540 | 1.60 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr17_+_663284 | 1.59 |
ENSDART00000172540
|
adssl1
|
adenylosuccinate synthase like 1 |
| chr23_+_27749014 | 1.57 |
ENSDART00000128833
|
rps26
|
ribosomal protein S26 |
| chr13_-_39365252 | 1.53 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr18_-_8638929 | 1.50 |
|
|
|
| chr2_+_44067425 | 1.48 |
ENSDART00000123673
|
si:ch211-195h23.3
|
si:ch211-195h23.3 |
| chr3_+_54327353 | 1.47 |
ENSDART00000127487
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr1_-_18110990 | 1.44 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr9_-_29056763 | 1.43 |
ENSDART00000060321
|
ENSDARG00000056704
|
ENSDARG00000056704 |
| chr25_-_31218193 | 1.42 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr11_+_21749658 | 1.41 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr13_-_49826330 | 1.41 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr9_-_14533551 | 1.40 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr2_-_41269732 | 1.40 |
ENSDART00000134756
|
hs6st1a
|
heparan sulfate 6-O-sulfotransferase 1a |
| chr10_-_8101868 | 1.40 |
ENSDART00000179549
|
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr7_+_55797053 | 1.40 |
ENSDART00000127046
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr15_+_28269289 | 1.39 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr17_-_31678247 | 1.36 |
ENSDART00000143090
|
lin52
|
lin-52 DREAM MuvB core complex component |
| chr12_+_17382681 | 1.32 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr10_-_42152404 | 1.32 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr10_-_36864268 | 1.32 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr1_-_9387290 | 1.32 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr17_+_10161999 | 1.31 |
ENSDART00000170274
|
foxa1
|
forkhead box A1 |
| chr8_+_47644421 | 1.30 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr1_-_10391257 | 1.28 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr8_+_7340538 | 1.27 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr14_+_47326080 | 1.27 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr21_-_39013447 | 1.24 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr19_+_20209561 | 1.24 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr21_-_4375313 | 1.23 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr6_+_45930323 | 1.20 |
ENSDART00000154896
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr24_-_33869817 | 1.20 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr25_-_32344881 | 1.19 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr1_+_33563354 | 1.19 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr14_-_45883839 | 1.19 |
|
|
|
| chr7_+_19560971 | 1.19 |
ENSDART00000044425
|
BX957278.1
|
ENSDARG00000010940 |
| chr25_+_30701751 | 1.19 |
|
|
|
| chr2_+_11902170 | 1.18 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr4_-_17420378 | 1.18 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr25_-_18855316 | 1.17 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr16_+_17069742 | 1.16 |
ENSDART00000132150
|
rpl18
|
ribosomal protein L18 |
| chr10_-_23939618 | 1.14 |
|
|
|
| chr8_+_52633069 | 1.14 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr3_-_34671480 | 1.14 |
ENSDART00000167355
|
nsfa
|
N-ethylmaleimide-sensitive factor a |
| chr3_+_31802145 | 1.14 |
ENSDART00000139644
|
lin7b
|
lin-7 homolog B (C. elegans) |
| chr2_-_28446615 | 1.14 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr10_+_9716807 | 1.14 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr23_-_4979524 | 1.13 |
ENSDART00000060714
|
atp6ap1a
|
ATPase, H+ transporting, lysosomal accessory protein 1a |
| chr9_+_907908 | 1.13 |
ENSDART00000144114
|
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr9_+_41278246 | 1.13 |
ENSDART00000141179
|
zgc:136439
|
zgc:136439 |
| chr19_-_6466494 | 1.13 |
ENSDART00000104950
|
atp1a3a
|
ATPase, Na+/K+ transporting, alpha 3a polypeptide |
| chr8_+_13465718 | 1.12 |
ENSDART00000034740
|
fut9d
|
fucosyltransferase 9d |
| chr12_-_10372055 | 1.11 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr16_+_46034246 | 1.10 |
ENSDART00000101753
|
mtmr11
|
myotubularin related protein 11 |
| chr5_-_1846709 | 1.10 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr3_-_11892052 | 1.09 |
|
|
|
| chr14_-_8975187 | 1.09 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr9_-_22324699 | 1.08 |
ENSDART00000175417
ENSDART00000101902 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr10_-_22834248 | 1.08 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr6_+_42821679 | 1.07 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr12_+_25684420 | 1.07 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr9_+_54692834 | 1.06 |
|
|
|
| chr9_+_32490721 | 1.06 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr14_+_905613 | 1.05 |
ENSDART00000111141
|
myoz3a
|
myozenin 3a |
| chr23_+_9285446 | 1.05 |
ENSDART00000033663
|
rps21
|
ribosomal protein S21 |
| chr22_-_23641813 | 1.05 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr19_-_44459390 | 1.04 |
ENSDART00000125168
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr23_-_4990023 | 1.04 |
ENSDART00000142699
|
taz
|
tafazzin |
| chr23_+_45020761 | 1.04 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr23_+_22729939 | 1.03 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr3_+_13040717 | 1.03 |
|
|
|
| chr7_-_60525749 | 1.03 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr11_-_42096921 | 1.03 |
|
|
|
| chr22_-_11712371 | 1.03 |
|
|
|
| chr24_-_11103952 | 1.01 |
|
|
|
| chr17_-_51032853 | 1.01 |
ENSDART00000130412
ENSDART00000123746 ENSDART00000162717 |
aqr
|
aquarius intron-binding spliceosomal factor |
| chr23_+_17856053 | 1.00 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr10_+_9764591 | 1.00 |
ENSDART00000091780
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr1_+_50395721 | 1.00 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr12_+_15624640 | 1.00 |
ENSDART00000079803
|
nmt1b
|
N-myristoyltransferase 1b |
| chr1_+_25662910 | 1.00 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr24_-_6617860 | 1.00 |
ENSDART00000166216
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr24_-_36840203 | 0.99 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr8_-_41194530 | 0.99 |
ENSDART00000173055
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr6_-_7281412 | 0.99 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr12_-_5628248 | 0.99 |
ENSDART00000105884
|
itga3b
|
integrin, alpha 3b |
| chr4_-_12726432 | 0.97 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr22_-_17652938 | 0.97 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr7_-_66653301 | 0.96 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr21_-_21477423 | 0.95 |
ENSDART00000164076
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr2_-_32521879 | 0.94 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr5_+_45822196 | 0.93 |
|
|
|
| chr9_+_219478 | 0.93 |
ENSDART00000164048
ENSDART00000161484 ENSDART00000171623 |
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr22_+_30060679 | 0.92 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr16_-_44575942 | 0.92 |
|
|
|
| chr3_+_13487523 | 0.92 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr17_+_1058137 | 0.92 |
|
|
|
| chr17_+_11904378 | 0.91 |
ENSDART00000150209
ENSDART00000110058 |
tfb2m
|
transcription factor B2, mitochondrial |
| chr3_+_24327586 | 0.91 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
| chr25_-_16547149 | 0.91 |
ENSDART00000104110
|
slc25a18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr5_+_50432868 | 0.90 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr9_-_35106146 | 0.89 |
ENSDART00000139608
ENSDART00000100728 ENSDART00000123005 |
upf3a
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr3_-_6633512 | 0.89 |
ENSDART00000165273
|
GGA3 (1 of many)
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr15_-_1225595 | 0.89 |
|
|
|
| chr5_-_45729685 | 0.89 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr21_-_4842528 | 0.89 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr2_+_37985514 | 0.89 |
ENSDART00000149224
|
apol1
|
apolipoprotein L, 1 |
| chr10_-_23130343 | 0.89 |
ENSDART00000148333
|
nle1
|
notchless homolog 1 (Drosophila) |
| chr12_+_46944395 | 0.88 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr16_+_53238110 | 0.88 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr22_+_24131239 | 0.88 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr19_-_9793494 | 0.88 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr3_-_57596004 | 0.87 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr16_+_22672315 | 0.86 |
ENSDART00000142241
|
she
|
Src homology 2 domain containing E |
| chr10_+_33926312 | 0.86 |
ENSDART00000174730
|
BX569785.2
|
ENSDARG00000106629 |
| chr24_-_32750010 | 0.86 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr4_-_16356071 | 0.86 |
ENSDART00000079521
|
kera
|
keratocan |
| chr16_-_42054448 | 0.85 |
ENSDART00000157645
|
caspa
|
caspase a |
| chr24_-_33617592 | 0.85 |
|
|
|
| chr6_-_54103765 | 0.85 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr4_+_17290988 | 0.85 |
ENSDART00000067005
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr1_+_9176411 | 0.85 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr12_+_19240407 | 0.85 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr5_+_36180981 | 0.84 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr16_-_44682957 | 0.84 |
|
|
|
| chr12_+_35018663 | 0.84 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr9_+_51733862 | 0.84 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr9_-_22371512 | 0.83 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr5_+_29920060 | 0.83 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr11_+_41324286 | 0.83 |
ENSDART00000055710
|
aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr4_+_12618256 | 0.83 |
ENSDART00000112860
ENSDART00000134362 |
lmo3
|
LIM domain only 3 |
| chr1_+_38099962 | 0.81 |
ENSDART00000166864
|
spcs3
|
signal peptidase complex subunit 3 |
| chr5_-_5338903 | 0.81 |
ENSDART00000112856
|
ENSDARG00000078362
|
ENSDARG00000078362 |
| chr7_-_44332679 | 0.80 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr16_-_4811237 | 0.80 |
ENSDART00000076955
|
cyp4t8
|
cytochrome P450, family 4, subfamily T, polypeptide 8 |
| chr3_-_39346621 | 0.80 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr3_-_2686533 | 0.79 |
|
|
|
| chr9_+_24198112 | 0.79 |
ENSDART00000148226
|
mlphb
|
melanophilin b |
| chr4_+_14958625 | 0.78 |
ENSDART00000002770
|
tspan33a
|
tetraspanin 33a |
| chr9_+_9524547 | 0.78 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr20_+_54463948 | 0.77 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr11_+_3940085 | 0.77 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr6_+_58549236 | 0.77 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr3_-_18261177 | 0.76 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr20_-_28739099 | 0.75 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr4_+_13569588 | 0.75 |
ENSDART00000136152
|
calua
|
calumenin a |
| chr9_+_3416525 | 0.75 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr1_+_44044807 | 0.75 |
ENSDART00000133926
ENSDART00000049701 |
p2rx3b
|
purinergic receptor P2X, ligand-gated ion channel, 3b |
| chr3_-_18605846 | 0.75 |
ENSDART00000145277
|
zgc:113333
|
zgc:113333 |
| chr2_-_54965656 | 0.75 |
|
|
|
| chr22_+_14077602 | 0.75 |
ENSDART00000146397
|
he2
|
hatching enzyme 2 |
| chr21_+_22386643 | 0.74 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr21_+_15737517 | 0.74 |
ENSDART00000151161
|
ENSDARG00000044755
|
ENSDARG00000044755 |
| chr11_-_16840339 | 0.73 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr20_-_28981879 | 0.73 |
ENSDART00000134564
ENSDART00000132127 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr15_-_18130992 | 0.73 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr17_-_35004687 | 0.72 |
|
|
|
| chr9_+_9524688 | 0.72 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr21_-_22672597 | 0.72 |
ENSDART00000140032
ENSDART00000136993 |
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr20_-_30997978 | 0.72 |
ENSDART00000153419
|
hebp2
|
heme binding protein 2 |
| chr14_+_31317100 | 0.72 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr20_+_7596461 | 0.71 |
ENSDART00000127975
ENSDART00000132481 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr3_-_32471212 | 0.71 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr9_+_9524768 | 0.71 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr5_+_38903966 | 0.71 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr8_-_14113377 | 0.70 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr3_+_52551672 | 0.70 |
ENSDART00000098826
|
lmx1al
|
LIM homeobox transcription factor 1, alpha-like |
| chr3_-_22082170 | 0.70 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr2_-_11879298 | 0.70 |
ENSDART00000136748
|
ENSDARG00000039203
|
ENSDARG00000039203 |
| chr25_-_14301843 | 0.70 |
ENSDART00000046934
|
coq9
|
coenzyme Q9 homolog (S. cerevisiae) |
| chr23_+_29962550 | 0.69 |
ENSDART00000149378
|
mxra8b
|
matrix-remodelling associated 8b |
| chr24_-_1308951 | 0.68 |
|
|
|
| chr12_+_40919018 | 0.68 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr6_+_1959688 | 0.67 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr17_+_19514507 | 0.67 |
|
|
|
| chr3_+_40667131 | 0.67 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.4 | 1.4 | GO:0055016 | hypochord development(GO:0055016) |
| 0.4 | 1.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 0.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 0.8 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 0.8 | GO:0090171 | chondrocyte morphogenesis(GO:0090171) |
| 0.3 | 3.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 0.5 | GO:0002526 | acute inflammatory response(GO:0002526) |
| 0.3 | 1.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 3.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.2 | 0.7 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 1.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.2 | 1.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.2 | 1.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 1.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 2.4 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 2.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.0 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.2 | 1.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.2 | 1.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 1.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 0.7 | GO:0061195 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.2 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 1.0 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.2 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.2 | 1.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.2 | 0.5 | GO:0070424 | peptidoglycan transport(GO:0015835) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.2 | 1.4 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.2 | 0.5 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.1 | 1.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 1.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.9 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.3 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.1 | 2.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.7 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 2.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.6 | GO:0032370 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.1 | 0.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.3 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.3 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.3 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.7 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.6 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 1.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 1.2 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 1.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 3.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.8 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 1.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 2.9 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.5 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.5 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 1.8 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 2.2 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 3.3 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.3 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 2.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 1.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) cell-substrate adhesion(GO:0031589) |
| 0.0 | 0.7 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.0 | 1.1 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.0 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.5 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 1.3 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.9 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.9 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.6 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 1.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 1.7 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.4 | 1.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.4 | 1.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 1.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 1.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 0.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 1.0 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 1.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 2.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 1.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.4 | 2.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 2.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 1.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.4 | 1.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.4 | 1.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 0.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 0.9 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.3 | 0.8 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.3 | 1.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.3 | 1.1 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 0.9 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 1.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.5 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 0.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.6 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.7 | GO:0004931 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 1.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 7.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.6 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0000215 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 5.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 3.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 1.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 2.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 1.2 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |