Project
DANIO-CODE
Navigation
Downloads
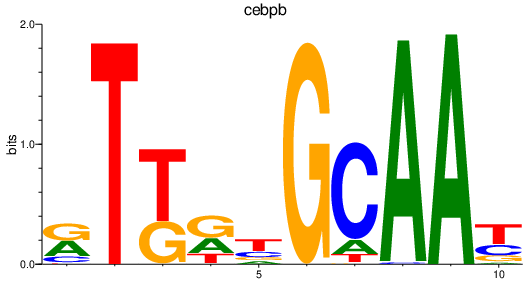
Results for cebpb
Z-value: 1.54
Transcription factors associated with cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpb
|
ENSDARG00000042725 | CCAAT enhancer binding protein beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpb | dr10_dc_chr8_-_28415750_28415816 | 0.54 | 3.2e-02 | Click! |
Activity profile of cebpb motif
Sorted Z-values of cebpb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_11765407 | 7.85 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr14_+_9115745 | 5.26 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr5_+_26195540 | 4.83 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr16_-_31670211 | 4.13 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr22_-_17570306 | 3.79 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr12_-_5085227 | 3.76 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr15_+_29153215 | 3.72 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr19_-_23037220 | 3.68 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr24_+_389982 | 3.46 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr14_+_20809272 | 3.32 |
ENSDART00000139865
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr1_+_25662652 | 3.19 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr14_-_30363703 | 3.18 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin-like extracellular matrix protein 2a |
| chr15_-_4322875 | 3.16 |
ENSDART00000173311
|
si:ch211-117a13.2
|
si:ch211-117a13.2 |
| chr13_-_18506153 | 3.14 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr13_-_39365252 | 3.14 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr5_+_42930239 | 3.04 |
ENSDART00000097618
|
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr2_+_7917776 | 2.96 |
ENSDART00000134178
ENSDART00000130781 |
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr5_+_50432868 | 2.96 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr21_-_39013447 | 2.95 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr7_+_63014869 | 2.92 |
ENSDART00000161436
|
pcdh7b
|
protocadherin 7b |
| chr11_+_23695123 | 2.90 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr2_-_30675594 | 2.89 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr8_+_15987710 | 2.88 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_-_5781358 | 2.87 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr1_+_49894978 | 2.81 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr5_-_43334777 | 2.80 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr5_-_62816208 | 2.80 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr16_-_21981065 | 2.73 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr25_+_7660590 | 2.69 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr25_+_30746445 | 2.69 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr12_+_25684420 | 2.64 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr2_+_36634309 | 2.64 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr7_-_60525749 | 2.60 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr2_-_28446615 | 2.57 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr11_-_42096921 | 2.51 |
|
|
|
| chr9_+_9524547 | 2.50 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr6_+_42478185 | 2.49 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr9_-_4890648 | 2.48 |
ENSDART00000167998
|
fmnl2a
|
formin-like 2a |
| chr18_+_618393 | 2.47 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr23_+_45020761 | 2.47 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr23_+_17856053 | 2.45 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr17_+_10161999 | 2.40 |
ENSDART00000170274
|
foxa1
|
forkhead box A1 |
| chr9_+_9524768 | 2.30 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr20_-_47521258 | 2.28 |
|
|
|
| chr6_+_7283773 | 2.28 |
ENSDART00000172285
|
arf3b
|
ADP-ribosylation factor 3b |
| chr9_+_48717490 | 2.24 |
ENSDART00000163353
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr5_+_45822196 | 2.23 |
|
|
|
| chr10_+_21850059 | 2.22 |
ENSDART00000164634
ENSDART00000172513 |
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr21_-_15833030 | 2.21 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr9_+_219478 | 2.21 |
ENSDART00000164048
ENSDART00000161484 ENSDART00000171623 |
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr1_+_25662910 | 2.14 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr10_-_23939618 | 2.11 |
|
|
|
| chr20_+_16843502 | 2.11 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr12_-_10372055 | 2.10 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr10_+_39291358 | 2.09 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr6_-_37444877 | 2.09 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr12_+_46944395 | 2.08 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr8_+_7340538 | 2.06 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr14_+_36545240 | 2.02 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr15_+_32853646 | 2.01 |
ENSDART00000167515
|
postnb
|
periostin, osteoblast specific factor b |
| chr13_+_28364942 | 2.00 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr7_+_21521123 | 1.99 |
ENSDART00000112531
ENSDART00000100936 |
tmem88b
|
transmembrane protein 88 b |
| chr7_-_66229124 | 1.96 |
ENSDART00000082604
|
galnt18b
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18b |
| chr9_-_35106146 | 1.96 |
ENSDART00000139608
ENSDART00000100728 ENSDART00000123005 |
upf3a
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| KN150702v1_+_177145 | 1.92 |
ENSDART00000166225
|
CABZ01076674.1
|
ENSDARG00000102620 |
| chr21_-_4842528 | 1.92 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr9_+_9524688 | 1.91 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr1_-_9387290 | 1.88 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr24_-_32750010 | 1.88 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr13_+_28365036 | 1.81 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr5_-_30015572 | 1.81 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr12_-_10001043 | 1.81 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr23_+_35996491 | 1.79 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr9_+_3416651 | 1.77 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr9_+_23854859 | 1.76 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr10_+_39140943 | 1.72 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr12_+_35018663 | 1.72 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr3_-_11892052 | 1.68 |
|
|
|
| chr6_+_58549236 | 1.67 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr3_-_39346621 | 1.67 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr6_+_16279737 | 1.65 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| KN150339v1_-_34304 | 1.64 |
|
|
|
| chr5_+_38903966 | 1.63 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr19_+_5073991 | 1.62 |
ENSDART00000151050
|
ENSDARG00000011076
|
ENSDARG00000011076 |
| chr3_+_23573114 | 1.61 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr22_+_15946773 | 1.60 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr21_-_9708608 | 1.59 |
|
|
|
| chr8_-_50159285 | 1.54 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr22_-_23641813 | 1.54 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr4_-_12726432 | 1.52 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr21_-_3301751 | 1.48 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr25_-_1989785 | 1.48 |
ENSDART00000156925
|
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr3_+_13487523 | 1.47 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr17_+_25500291 | 1.47 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr12_-_36415239 | 1.45 |
|
|
|
| chr24_-_11103952 | 1.45 |
|
|
|
| chr10_-_36864268 | 1.45 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr16_+_21090083 | 1.44 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr7_-_25816549 | 1.43 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr3_-_45420882 | 1.43 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr21_-_91068 | 1.42 |
|
|
|
| chr8_+_46378250 | 1.39 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr18_+_26916897 | 1.37 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr3_-_13942505 | 1.37 |
|
|
|
| chr8_+_46378706 | 1.36 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr22_+_16293900 | 1.36 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr23_+_11350727 | 1.36 |
|
|
|
| chr23_+_25378921 | 1.34 |
ENSDART00000143291
|
RPL41
|
ribosomal protein L41 |
| chr20_-_52459338 | 1.34 |
ENSDART00000135463
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr9_+_3416525 | 1.33 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr4_+_12618485 | 1.32 |
ENSDART00000112860
|
lmo3
|
LIM domain only 3 |
| chr22_-_17652938 | 1.31 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr17_+_52736535 | 1.28 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr24_-_36840203 | 1.27 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr23_+_22729939 | 1.26 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr25_-_17142578 | 1.25 |
ENSDART00000152106
|
ccnd2a
|
cyclin D2, a |
| chr12_+_25684362 | 1.23 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr3_+_40667131 | 1.22 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr6_-_37444807 | 1.22 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr21_-_8304754 | 1.22 |
ENSDART00000055328
|
nek6
|
NIMA-related kinase 6 |
| chr19_-_9793494 | 1.21 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr9_+_32490721 | 1.20 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr23_-_19053587 | 1.19 |
|
|
|
| chr15_-_3120500 | 1.18 |
|
|
|
| chr2_+_20814888 | 1.18 |
ENSDART00000170793
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr2_+_11245538 | 1.17 |
ENSDART00000138737
ENSDART00000081058 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr6_-_14162846 | 1.17 |
ENSDART00000100762
|
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr21_-_11561855 | 1.16 |
ENSDART00000162426
|
cast
|
calpastatin |
| chr2_+_27005381 | 1.16 |
|
|
|
| chr13_-_18064929 | 1.15 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr6_-_3413472 | 1.15 |
ENSDART00000026693
|
dmbx1b
|
diencephalon/mesencephalon homeobox 1b |
| chr17_-_43475725 | 1.15 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr13_+_40311601 | 1.14 |
ENSDART00000148510
|
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr24_-_1308951 | 1.12 |
|
|
|
| chr21_+_4375508 | 1.12 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr16_-_16793728 | 1.11 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr10_-_7427767 | 1.07 |
ENSDART00000167963
ENSDART00000162021 |
nrg1
|
neuregulin 1 |
| chr11_+_24479085 | 1.05 |
ENSDART00000145217
|
ENSDARG00000070571
|
ENSDARG00000070571 |
| chr8_-_37011436 | 1.05 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr17_-_35004687 | 1.05 |
|
|
|
| chr15_-_17074348 | 1.03 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr6_-_34237257 | 1.02 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr7_+_10346939 | 1.01 |
|
|
|
| chr25_+_1773157 | 1.01 |
|
|
|
| chr6_+_54134129 | 1.01 |
ENSDART00000156554
|
hmga1b
|
high mobility group AT-hook 1b |
| chr20_-_28739099 | 1.00 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr14_-_46646451 | 0.99 |
|
|
|
| chr23_-_21288972 | 0.99 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr21_+_18295124 | 0.99 |
ENSDART00000167511
|
ENSDARG00000104101
|
ENSDARG00000104101 |
| chr17_+_10345498 | 0.98 |
ENSDART00000081659
ENSDART00000169120 ENSDART00000170970 ENSDART00000140391 |
tyro3
|
TYRO3 protein tyrosine kinase |
| chr17_+_52736844 | 0.97 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr17_+_11904378 | 0.97 |
ENSDART00000150209
ENSDART00000110058 |
tfb2m
|
transcription factor B2, mitochondrial |
| chr21_+_29190291 | 0.97 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr4_-_885413 | 0.96 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr16_+_33189439 | 0.96 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr23_+_2665 | 0.96 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr4_-_22617898 | 0.95 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr13_-_45018897 | 0.94 |
|
|
|
| chr15_-_17074107 | 0.94 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr5_+_22204129 | 0.94 |
|
|
|
| chr19_-_46058895 | 0.94 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr9_-_7260431 | 0.90 |
ENSDART00000142726
|
creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr8_+_16954190 | 0.90 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr21_-_11554253 | 0.90 |
ENSDART00000133443
|
cast
|
calpastatin |
| chr14_-_46646747 | 0.89 |
|
|
|
| chr1_-_24486146 | 0.88 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr2_+_2295529 | 0.88 |
ENSDART00000127286
|
si:ch73-140j24.4
|
si:ch73-140j24.4 |
| chr20_+_29307142 | 0.87 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_+_21928819 | 0.86 |
ENSDART00000154622
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr24_-_14432269 | 0.85 |
ENSDART00000140624
|
pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr7_+_65532871 | 0.85 |
ENSDART00000082740
|
CT573494.2
|
ENSDARG00000097673 |
| chr9_+_23854967 | 0.84 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr10_-_42155708 | 0.83 |
|
|
|
| chr15_+_24277664 | 0.83 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr3_+_13040717 | 0.82 |
|
|
|
| chr4_-_19705018 | 0.81 |
ENSDART00000100974
|
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr22_+_15412270 | 0.81 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr13_+_41791642 | 0.77 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr9_-_30453625 | 0.73 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr14_+_50011198 | 0.72 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr10_-_42155790 | 0.71 |
|
|
|
| chr20_+_29307039 | 0.69 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr21_+_4345150 | 0.69 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr18_-_20880226 | 0.68 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr10_+_45401582 | 0.68 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr7_-_52692386 | 0.65 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr8_-_46378116 | 0.64 |
ENSDART00000136602
|
qars
|
glutaminyl-tRNA synthetase |
| chr6_+_28215039 | 0.63 |
ENSDART00000104394
|
smx5
|
smx5 |
| chr9_-_34690993 | 0.62 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr22_-_18466781 | 0.61 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr14_-_1080574 | 0.61 |
ENSDART00000169090
ENSDART00000158905 ENSDART00000159907 |
ENSDARG00000098694
|
ENSDARG00000098694 |
| chr9_-_23577201 | 0.60 |
ENSDART00000141524
|
CR392363.2
|
ENSDARG00000095527 |
| chr17_-_17928974 | 0.60 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr11_+_36984616 | 0.59 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr14_-_46646556 | 0.59 |
|
|
|
| chr7_-_25816498 | 0.58 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr23_-_35904414 | 0.58 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr21_+_21706999 | 0.56 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr9_-_34691159 | 0.55 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr24_+_16839260 | 0.54 |
ENSDART00000175812
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 1.0 | 3.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 1.0 | 3.8 | GO:0043586 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.9 | 2.8 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.8 | 5.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.8 | 3.3 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.8 | 3.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.7 | 2.9 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.7 | 2.0 | GO:0006581 | response to amphetamine(GO:0001975) acetylcholine catabolic process(GO:0006581) response to amine(GO:0014075) |
| 0.6 | 3.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 1.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 3.8 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.4 | 1.8 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.4 | 2.1 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 2.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.4 | 1.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.4 | 2.2 | GO:0070293 | renal absorption(GO:0070293) |
| 0.4 | 4.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.4 | 2.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.4 | 1.1 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.3 | 1.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 1.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 1.0 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.3 | 1.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 2.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 2.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 2.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.3 | 3.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 2.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 5.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.2 | 2.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 1.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 2.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 2.4 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.2 | 1.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 3.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 2.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.2 | 2.0 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 3.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 2.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 2.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.5 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 1.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0071295 | cellular response to nutrient(GO:0031670) vitamin D3 metabolic process(GO:0070640) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 1.8 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 1.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 3.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 3.0 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 2.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.6 | GO:0032944 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.1 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.9 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 3.1 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 1.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 1.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 3.3 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 1.0 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 1.5 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 2.0 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 2.2 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 2.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.4 | GO:0033333 | fin development(GO:0033333) |
| 0.0 | 2.2 | GO:0030900 | forebrain development(GO:0030900) |
| 0.0 | 0.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.6 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.8 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 3.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.4 | 2.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.4 | 1.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 2.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 3.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.5 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.2 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 5.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 2.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.9 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 25.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 8.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 3.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.1 | 5.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.0 | 3.0 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.9 | 3.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.8 | 3.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.8 | 3.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.7 | 2.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.7 | 2.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 2.7 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.7 | 2.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.6 | 2.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.6 | 4.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 1.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 3.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.4 | 5.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 2.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 1.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.3 | 3.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 3.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 6.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.3 | 1.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 3.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 2.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 1.0 | GO:0004937 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 1.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 1.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 7.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.2 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 5.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 3.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.3 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.8 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 6.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 9.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.2 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 2.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 2.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 2.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 2.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 5.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 1.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 2.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 6.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 2.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 2.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 6.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.6 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 3.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 2.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |