Project
DANIO-CODE
Navigation
Downloads
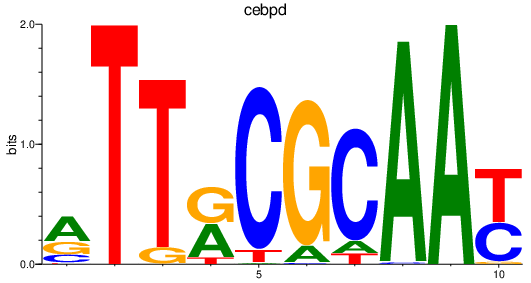
Results for cebpd
Z-value: 0.45
Transcription factors associated with cebpd
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpd
|
ENSDARG00000087303 | CCAAT enhancer binding protein delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpd | dr10_dc_chr24_+_35676505_35676557 | 0.53 | 3.5e-02 | Click! |
Activity profile of cebpd motif
Sorted Z-values of cebpd motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpd
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_11765407 | 1.20 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr16_+_24069711 | 1.14 |
ENSDART00000142869
|
apoc2
|
apolipoprotein C-II |
| chr11_+_23695123 | 1.01 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr19_-_23037220 | 0.92 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr1_-_9387290 | 0.92 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr5_+_26195540 | 0.87 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr7_+_63014869 | 0.86 |
ENSDART00000161436
|
pcdh7b
|
protocadherin 7b |
| chr4_-_17420378 | 0.84 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr10_-_23939618 | 0.83 |
|
|
|
| chr9_+_3416651 | 0.82 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr16_+_46145286 | 0.80 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_-_24982964 | 0.78 |
ENSDART00000031665
ENSDART00000161043 |
anxa1a
|
annexin A1a |
| chr6_-_11544518 | 0.75 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr23_+_45020761 | 0.72 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr24_-_32750010 | 0.71 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr18_+_618393 | 0.69 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr24_+_389982 | 0.69 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr15_-_4322875 | 0.68 |
ENSDART00000173311
|
si:ch211-117a13.2
|
si:ch211-117a13.2 |
| chr22_-_17570306 | 0.65 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr17_-_43475725 | 0.63 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr12_+_35018663 | 0.63 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr4_-_5294280 | 0.62 |
ENSDART00000178921
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr9_+_219478 | 0.62 |
ENSDART00000164048
ENSDART00000161484 ENSDART00000171623 |
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr8_+_7340538 | 0.61 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr16_-_31670211 | 0.60 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr5_-_43334777 | 0.57 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr5_-_62816208 | 0.56 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr11_-_44724371 | 0.55 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr12_+_25684420 | 0.54 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr6_-_48095609 | 0.52 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr8_+_4311821 | 0.51 |
ENSDART00000015214
|
pptc7a
|
PTC7 protein phosphatase homolog a |
| KN150702v1_+_177145 | 0.51 |
ENSDART00000166225
|
CABZ01076674.1
|
ENSDARG00000102620 |
| chr7_-_66229124 | 0.48 |
ENSDART00000082604
|
galnt18b
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18b |
| chr7_-_42972804 | 0.47 |
ENSDART00000054552
|
cdh8
|
cadherin 8 |
| chr5_+_50432868 | 0.46 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr22_+_24617667 | 0.46 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr9_+_3416525 | 0.46 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr16_-_17164459 | 0.45 |
ENSDART00000178443
|
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr1_+_25378105 | 0.44 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr16_+_5470544 | 0.44 |
|
|
|
| chr20_-_37733350 | 0.44 |
|
|
|
| chr22_+_15946773 | 0.43 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr25_+_7660590 | 0.43 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr7_+_39147543 | 0.43 |
ENSDART00000165294
|
CT030188.1
|
ENSDARG00000099385 |
| chr25_+_33924376 | 0.42 |
|
|
|
| chr9_+_48717490 | 0.42 |
ENSDART00000163353
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr16_+_17705704 | 0.41 |
|
|
|
| chr5_+_31562242 | 0.40 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr2_+_2295529 | 0.40 |
ENSDART00000127286
|
si:ch73-140j24.4
|
si:ch73-140j24.4 |
| chr17_-_10682357 | 0.40 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr25_-_17142578 | 0.38 |
ENSDART00000152106
|
ccnd2a
|
cyclin D2, a |
| chr24_-_36840203 | 0.38 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr18_-_20880226 | 0.37 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr20_-_45757288 | 0.37 |
ENSDART00000124582
ENSDART00000100290 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr23_-_5080531 | 0.35 |
|
|
|
| chr24_-_39884167 | 0.35 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr21_+_29190291 | 0.34 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr6_-_14162846 | 0.34 |
ENSDART00000100762
|
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr2_+_29862841 | 0.34 |
ENSDART00000135918
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr19_-_44459390 | 0.34 |
ENSDART00000125168
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr6_-_7937201 | 0.34 |
|
|
|
| chr16_+_44333789 | 0.33 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr10_-_42155790 | 0.33 |
|
|
|
| chr19_-_15288270 | 0.33 |
ENSDART00000151451
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr9_-_35260503 | 0.31 |
ENSDART00000125536
|
appb
|
amyloid beta (A4) precursor protein b |
| chr12_+_25684362 | 0.31 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr21_-_39013447 | 0.31 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr4_-_5293599 | 0.31 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr12_-_36415239 | 0.30 |
|
|
|
| chr24_+_38783264 | 0.29 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr21_+_4345150 | 0.29 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr7_-_52692386 | 0.29 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr16_+_21090083 | 0.28 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr10_+_38582701 | 0.28 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr1_-_25377787 | 0.27 |
|
|
|
| chr12_+_34852049 | 0.27 |
ENSDART00000027034
|
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr21_-_91068 | 0.27 |
|
|
|
| chr17_-_5918853 | 0.26 |
ENSDART00000058890
ENSDART00000171084 |
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr16_+_17706003 | 0.26 |
|
|
|
| chr10_+_9762039 | 0.26 |
|
|
|
| chr15_+_31956131 | 0.25 |
|
|
|
| chr2_-_54662966 | 0.25 |
ENSDART00000162403
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr18_+_26916897 | 0.25 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr17_+_10161999 | 0.25 |
ENSDART00000170274
|
foxa1
|
forkhead box A1 |
| chr10_+_39140943 | 0.24 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr12_+_46944395 | 0.24 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr13_+_40311601 | 0.24 |
ENSDART00000148510
|
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr16_+_4288700 | 0.24 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr9_+_9524768 | 0.24 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr25_-_16890563 | 0.24 |
ENSDART00000073419
|
ccnd2a
|
cyclin D2, a |
| chr25_-_211879 | 0.23 |
ENSDART00000021614
|
srpk2
|
SRSF protein kinase 2 |
| chr23_+_11350727 | 0.22 |
|
|
|
| chr8_-_37011436 | 0.22 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr2_+_11245538 | 0.21 |
ENSDART00000138737
ENSDART00000081058 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr9_+_41910191 | 0.21 |
|
|
|
| chr7_+_67510360 | 0.21 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr6_+_42478185 | 0.20 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr12_-_20251183 | 0.20 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr5_+_52198613 | 0.19 |
ENSDART00000162459
|
scarb2a
|
scavenger receptor class B, member 2a |
| chr22_-_18466781 | 0.19 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr8_-_15071283 | 0.19 |
|
|
|
| chr3_+_23573114 | 0.18 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr9_-_12063411 | 0.18 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr5_-_28888658 | 0.18 |
|
|
|
| chr5_-_28888704 | 0.18 |
|
|
|
| chr9_-_7260431 | 0.17 |
ENSDART00000142726
|
creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr22_-_12723704 | 0.17 |
ENSDART00000080741
|
plcd4a
|
phospholipase C, delta 4a |
| chr20_-_27482535 | 0.16 |
ENSDART00000162430
|
serpina10a
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a |
| chr13_+_28574593 | 0.16 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr25_-_1989785 | 0.16 |
ENSDART00000156925
|
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr14_-_38469172 | 0.15 |
ENSDART00000159159
|
ENSDARG00000100564
|
ENSDARG00000100564 |
| chr16_+_5470419 | 0.14 |
|
|
|
| chr2_-_10300329 | 0.14 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr11_+_36984616 | 0.12 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr10_-_42155708 | 0.12 |
|
|
|
| chr9_+_32490721 | 0.12 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr4_-_22617898 | 0.10 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr16_-_53008678 | 0.10 |
|
|
|
| chr19_+_16160028 | 0.10 |
ENSDART00000144376
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr7_-_22519196 | 0.09 |
ENSDART00000129919
|
ENSDARG00000091111
|
ENSDARG00000091111 |
| chr10_+_45401582 | 0.09 |
ENSDART00000159797
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr21_-_11877044 | 0.08 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr24_-_38782898 | 0.08 |
ENSDART00000155542
|
FP102170.1
|
ENSDARG00000097123 |
| chr20_+_39112357 | 0.08 |
|
|
|
| chr22_-_16351483 | 0.07 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr7_-_39480446 | 0.07 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr7_-_64889963 | 0.07 |
ENSDART00000110614
ENSDART00000098277 ENSDART00000171493 |
tmem41b
|
transmembrane protein 41B |
| chr2_+_50126491 | 0.07 |
ENSDART00000144060
|
rpl37
|
ribosomal protein L37 |
| chr2_-_50126390 | 0.06 |
ENSDART00000083690
|
blvra
|
biliverdin reductase A |
| chr15_+_20607816 | 0.05 |
ENSDART00000169941
|
sgsm2
|
small G protein signaling modulator 2 |
| chr23_-_35904414 | 0.05 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr12_-_26291790 | 0.04 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr24_+_10942742 | 0.04 |
ENSDART00000136614
|
zfand1
|
zinc finger, AN1-type domain 1 |
| chr3_-_30948297 | 0.04 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr5_+_18837448 | 0.04 |
|
|
|
| chr9_+_24255064 | 0.03 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr25_+_3168376 | 0.03 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr22_-_16351555 | 0.02 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr2_+_55460609 | 0.02 |
ENSDART00000016143
|
ENSDARG00000001817
|
ENSDARG00000001817 |
| chr14_+_7596207 | 0.02 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr15_-_29415309 | 0.02 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr1_+_25110899 | 0.01 |
ENSDART00000129471
|
gucy1b3
|
guanylate cyclase 1, soluble, beta 3 |
| chr12_+_13167615 | 0.01 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr10_-_38582881 | 0.01 |
ENSDART00000157222
|
BX000451.1
|
ENSDARG00000097896 |
| chr21_+_9904707 | 0.01 |
ENSDART00000171579
|
herc7
|
hect domain and RLD 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1902869 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.3 | 0.8 | GO:0045064 | positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) negative regulation of phospholipase activity(GO:0010519) positive regulation of vesicle fusion(GO:0031340) type 2 immune response(GO:0042092) regulation of fatty acid biosynthetic process(GO:0042304) T-helper 2 cell differentiation(GO:0045064) leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.3 | 1.0 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.8 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.4 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.5 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.3 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 1.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.4 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.7 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.2 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.7 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.2 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.4 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.4 | GO:0002697 | regulation of immune effector process(GO:0002697) |
| 0.0 | 0.2 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.7 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.4 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.8 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0004937 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.5 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |