Project
DANIO-CODE
Navigation
Downloads
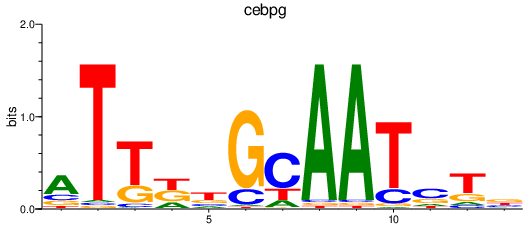
Results for cebpg
Z-value: 0.50
Transcription factors associated with cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpg
|
ENSDARG00000036073 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpg | dr10_dc_chr7_+_37818365_37818503 | 0.52 | 3.8e-02 | Click! |
Activity profile of cebpg motif
Sorted Z-values of cebpg motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpg
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_20819101 | 0.58 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr6_+_10098305 | 0.57 |
ENSDART00000151477
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr4_-_14916491 | 0.45 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr15_+_6462447 | 0.44 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr15_+_6462607 | 0.43 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr8_+_28715451 | 0.43 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr21_+_43183522 | 0.43 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr17_-_11904068 | 0.42 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr9_+_32262914 | 0.42 |
ENSDART00000110204
|
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr20_+_41351373 | 0.41 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| chr10_-_42152265 | 0.37 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr22_+_26840517 | 0.37 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr9_+_34423285 | 0.33 |
ENSDART00000174944
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr17_+_12788541 | 0.32 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr5_+_17508128 | 0.32 |
ENSDART00000048859
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr19_-_2928582 | 0.31 |
ENSDART00000109130
|
recql4
|
RecQ helicase-like 4 |
| chr5_-_69171508 | 0.31 |
ENSDART00000127782
|
ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr23_-_34017934 | 0.31 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr7_+_37818365 | 0.30 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr9_-_53522365 | 0.29 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr3_-_39163141 | 0.27 |
ENSDART00000102674
ENSDART00000167070 |
plcd3a
|
phospholipase C, delta 3a |
| chr8_+_28715581 | 0.27 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr19_-_44459352 | 0.27 |
ENSDART00000151084
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr22_+_16070849 | 0.26 |
ENSDART00000165216
|
dph5
|
diphthamide biosynthesis 5 |
| chr13_-_3805904 | 0.26 |
ENSDART00000017052
|
ncoa4
|
nuclear receptor coactivator 4 |
| chr18_-_37271104 | 0.26 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr5_-_14783255 | 0.25 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr3_+_33308831 | 0.25 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr19_-_20819028 | 0.25 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr4_-_14916416 | 0.25 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr7_+_26730804 | 0.24 |
|
|
|
| chr3_-_39163225 | 0.24 |
ENSDART00000102674
ENSDART00000167070 |
plcd3a
|
phospholipase C, delta 3a |
| chr9_-_53522524 | 0.24 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr9_+_19812730 | 0.24 |
ENSDART00000110009
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr14_-_26093969 | 0.23 |
ENSDART00000037999
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr13_-_25711537 | 0.22 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr7_+_37818298 | 0.22 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr16_-_43137552 | 0.21 |
ENSDART00000155445
|
si:dkey-7j14.6
|
si:dkey-7j14.6 |
| chr18_-_35485549 | 0.21 |
ENSDART00000141023
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr16_-_7911453 | 0.21 |
ENSDART00000108653
|
tcaim
|
T-cell activation inhibitor, mitochondrial |
| chr17_+_18012036 | 0.21 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr21_-_30387895 | 0.21 |
|
|
|
| chr1_+_8010026 | 0.21 |
ENSDART00000152142
|
zgc:77849
|
zgc:77849 |
| chr5_+_63596281 | 0.20 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr20_-_16366248 | 0.20 |
|
|
|
| chr17_+_11904378 | 0.19 |
ENSDART00000150209
ENSDART00000110058 |
tfb2m
|
transcription factor B2, mitochondrial |
| chr15_-_45378317 | 0.19 |
ENSDART00000179432
|
CABZ01067262.1
|
ENSDARG00000107582 |
| chr1_+_40428827 | 0.19 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr6_+_8916643 | 0.18 |
|
|
|
| chr10_+_6494926 | 0.18 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr3_+_25719002 | 0.17 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr19_-_35392661 | 0.17 |
ENSDART00000103665
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr19_-_20819057 | 0.17 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr8_-_18547147 | 0.17 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr17_+_28653205 | 0.16 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr6_+_28027815 | 0.16 |
ENSDART00000123324
|
sap130a
|
Sin3A-associated protein a |
| chr5_+_58360376 | 0.16 |
ENSDART00000110182
|
rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr20_+_31381577 | 0.16 |
|
|
|
| chr15_-_34106827 | 0.14 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr20_-_16366304 | 0.14 |
|
|
|
| chr4_-_765957 | 0.14 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr18_-_5656654 | 0.14 |
ENSDART00000142945
ENSDART00000063024 |
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr3_-_26060047 | 0.14 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr5_-_69171051 | 0.14 |
ENSDART00000127782
|
ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr1_+_40428722 | 0.14 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr9_+_2472639 | 0.14 |
ENSDART00000147034
|
gpr155a
|
G protein-coupled receptor 155a |
| chr20_+_41351418 | 0.14 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| chr25_-_22954929 | 0.13 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr13_-_14798134 | 0.13 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr25_+_34306049 | 0.13 |
ENSDART00000087410
|
adat1
|
adenosine deaminase, tRNA-specific 1 |
| chr14_-_26093882 | 0.13 |
ENSDART00000170614
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
| chr24_+_32610430 | 0.13 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr25_-_26309873 | 0.13 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr21_+_43183835 | 0.12 |
ENSDART00000175107
|
aff4
|
AF4/FMR2 family, member 4 |
| chr22_-_16291512 | 0.12 |
ENSDART00000163664
|
trmt13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr5_+_26195540 | 0.12 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr2_+_30800532 | 0.12 |
|
|
|
| chr12_-_9049697 | 0.12 |
ENSDART00000022163
ENSDART00000152793 |
exoc6
|
exocyst complex component 6 |
| chr17_-_24420654 | 0.11 |
|
|
|
| chr3_+_40667131 | 0.11 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr10_-_7827244 | 0.11 |
ENSDART00000111058
|
mpx
|
myeloid-specific peroxidase |
| chr3_+_23573053 | 0.11 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr6_+_37551455 | 0.11 |
ENSDART00000160960
ENSDART00000039934 ENSDART00000136585 |
slc9a7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr22_-_18466781 | 0.11 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr22_-_18466730 | 0.10 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr24_+_32610480 | 0.10 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr17_+_28653236 | 0.10 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr14_-_14400068 | 0.10 |
ENSDART00000170269
|
ogt.2
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 2 |
| chr7_-_67741663 | 0.10 |
|
|
|
| chr8_+_31247312 | 0.10 |
ENSDART00000174921
|
AL929453.2
|
ENSDARG00000108092 |
| chr17_-_20198477 | 0.10 |
ENSDART00000063523
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr1_+_45817971 | 0.10 |
|
|
|
| chr14_-_31748733 | 0.09 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr24_-_38757086 | 0.09 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr13_+_11307037 | 0.09 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_+_30941618 | 0.09 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr3_+_23573114 | 0.09 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr21_+_43183570 | 0.09 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr13_+_11306998 | 0.09 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_-_26060098 | 0.09 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr9_-_32489944 | 0.08 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr7_+_24983985 | 0.08 |
|
|
|
| chr6_-_39346614 | 0.08 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr16_-_42054448 | 0.08 |
ENSDART00000157645
|
caspa
|
caspase a |
| chr18_+_5657314 | 0.08 |
ENSDART00000007797
|
slc30a4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr14_-_46564205 | 0.08 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr20_+_40560443 | 0.08 |
ENSDART00000153119
|
serinc1
|
serine incorporator 1 |
| chr15_+_34106908 | 0.08 |
|
|
|
| chr19_-_10997025 | 0.08 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr9_-_4890587 | 0.07 |
ENSDART00000167998
|
fmnl2a
|
formin-like 2a |
| chr10_+_23130414 | 0.07 |
ENSDART00000022170
|
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr7_-_18816199 | 0.07 |
ENSDART00000108787
|
ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr17_+_51654782 | 0.07 |
ENSDART00000149039
|
odc1
|
ornithine decarboxylase 1 |
| chr20_-_52459338 | 0.07 |
ENSDART00000135463
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr2_+_30965711 | 0.06 |
|
|
|
| chr2_-_9898419 | 0.06 |
ENSDART00000081214
ENSDART00000171529 |
selt1a
|
selenoprotein T, 1a |
| chr18_-_37270932 | 0.06 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr23_-_4979524 | 0.06 |
ENSDART00000060714
|
atp6ap1a
|
ATPase, H+ transporting, lysosomal accessory protein 1a |
| chr3_+_33309011 | 0.06 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr2_-_9898112 | 0.05 |
ENSDART00000165712
|
selt1a
|
selenoprotein T, 1a |
| chr14_-_46646688 | 0.05 |
|
|
|
| chr23_+_2256243 | 0.05 |
ENSDART00000126768
|
cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr5_-_31474310 | 0.05 |
ENSDART00000112342
|
arpc5lb
|
actin related protein 2/3 complex, subunit 5-like, b |
| chr24_-_33370476 | 0.05 |
ENSDART00000155429
|
zgc:195173
|
zgc:195173 |
| chr13_-_24130160 | 0.05 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr11_-_34521855 | 0.04 |
ENSDART00000039847
ENSDART00000135725 |
chchd4a
|
coiled-coil-helix-coiled-coil-helix domain containing 4a |
| chr24_-_32750010 | 0.04 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr5_-_28888658 | 0.04 |
|
|
|
| chr9_+_42805274 | 0.04 |
ENSDART00000138133
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr16_-_31670211 | 0.04 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr13_-_25066650 | 0.03 |
|
|
|
| chr3_+_26023021 | 0.03 |
|
|
|
| chr23_-_30772173 | 0.03 |
ENSDART00000138841
|
CR456642.1
|
ENSDARG00000092307 |
| chr10_+_33956052 | 0.03 |
|
|
|
| chr23_-_34018069 | 0.03 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr17_-_11214364 | 0.03 |
ENSDART00000155976
|
FP102895.1
|
ENSDARG00000097935 |
| chr11_+_43127567 | 0.03 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr9_+_2472749 | 0.03 |
ENSDART00000147034
|
gpr155a
|
G protein-coupled receptor 155a |
| chr3_+_40667088 | 0.03 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr5_+_3965645 | 0.03 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr25_-_26292712 | 0.03 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr16_-_31386496 | 0.03 |
|
|
|
| chr3_-_15905495 | 0.02 |
ENSDART00000023859
|
atp6v0ca
|
ATPase, H+ transporting, lysosomal, V0 subunit ca |
| chr20_+_41351477 | 0.02 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| chr14_+_41039161 | 0.02 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr18_-_15582896 | 0.02 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| KN150339v1_-_34277 | 0.02 |
|
|
|
| chr12_+_27038210 | 0.02 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr8_+_15987710 | 0.02 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr21_-_22672597 | 0.01 |
ENSDART00000140032
ENSDART00000136993 |
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr15_+_9351211 | 0.01 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr9_-_40863087 | 0.01 |
ENSDART00000135134
|
bard1
|
BRCA1 associated RING domain 1 |
| chr21_+_4375508 | 0.01 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr1_+_44098980 | 0.01 |
ENSDART00000073712
|
tmem187
|
transmembrane protein 187 |
| chr15_-_21078312 | 0.01 |
ENSDART00000144991
|
ENSDARG00000093199
|
ENSDARG00000093199 |
| chr8_-_18547116 | 0.00 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr10_-_28307019 | 0.00 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr9_+_426850 | 0.00 |
ENSDART00000164310
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr13_+_11306913 | 0.00 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr25_-_2570269 | 0.00 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr17_+_777154 | 0.00 |
ENSDART00000161446
|
si:ch211-193k8.2
|
si:ch211-193k8.2 |
| chr1_+_55074914 | 0.00 |
ENSDART00000134770
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.9 | GO:0097324 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 0.3 | GO:0010984 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.1 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.5 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.1 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0010971 | regulation of mitotic cell cycle, embryonic(GO:0009794) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic cell cycle, embryonic(GO:0045448) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 0.2 | GO:0008942 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.1 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |