Project
DANIO-CODE
Navigation
Downloads
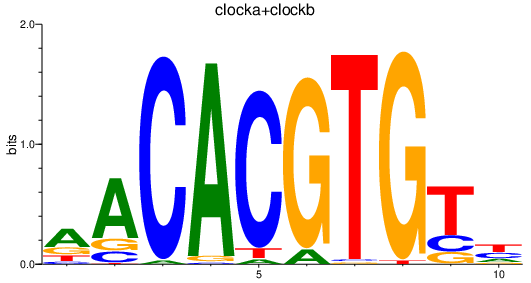
Results for clocka+clockb
Z-value: 2.12
Transcription factors associated with clocka+clockb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
clockb
|
ENSDARG00000003631 | clock circadian regulator b |
|
clocka
|
ENSDARG00000011703 | clock circadian regulator a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| clockb | dr10_dc_chr1_+_18739982_18740119 | -0.62 | 9.9e-03 | Click! |
Activity profile of clocka+clockb motif
Sorted Z-values of clocka+clockb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of clocka+clockb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_22065599 | 8.67 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr12_+_13053552 | 6.87 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr5_-_15971338 | 5.79 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr16_-_34240818 | 5.64 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr22_-_20670164 | 4.65 |
ENSDART00000169077
|
org
|
oogenesis-related gene |
| chr9_+_25109716 | 3.95 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_-_17163526 | 3.80 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr13_-_11888944 | 3.78 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr18_+_14725283 | 3.72 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr10_+_14984467 | 3.51 |
|
|
|
| chr22_+_17235696 | 3.51 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr16_-_31834830 | 3.49 |
|
|
|
| chr15_-_14102102 | 3.46 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr15_+_5125138 | 3.42 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr9_-_11616001 | 3.37 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr5_+_24039736 | 3.35 |
ENSDART00000175389
|
atp6v0a2b
|
ATPase, H+ transporting, lysosomal V0 subunit a2b |
| chr13_-_23626146 | 3.34 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr19_+_43984165 | 3.32 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr9_+_20970156 | 3.30 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr25_+_22474794 | 3.26 |
ENSDART00000152096
|
stra6
|
stimulated by retinoic acid 6 |
| chr13_+_35799681 | 3.21 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr13_-_11888896 | 3.20 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr14_-_21779536 | 3.15 |
|
|
|
| chr25_+_21001126 | 3.12 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr16_+_25344257 | 3.09 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr24_+_36429525 | 3.08 |
ENSDART00000062722
|
pus3
|
pseudouridylate synthase 3 |
| chr20_+_42640466 | 3.04 |
ENSDART00000134066
|
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr24_+_34183462 | 3.02 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr3_-_58094618 | 2.92 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr16_-_21734502 | 2.91 |
ENSDART00000179125
|
CABZ01009710.1
|
ENSDARG00000106589 |
| chr1_+_13768098 | 2.89 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr22_+_818795 | 2.88 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr11_+_44678488 | 2.88 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr6_+_3556296 | 2.87 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr1_+_5437327 | 2.84 |
ENSDART00000092324
|
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr16_-_38383875 | 2.84 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr5_+_37144525 | 2.78 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr7_-_38581532 | 2.74 |
|
|
|
| chr1_+_49171052 | 2.74 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr25_-_8548041 | 2.71 |
ENSDART00000155280
|
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr5_-_1604221 | 2.71 |
ENSDART00000055878
|
rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr10_+_22065010 | 2.71 |
ENSDART00000133304
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr8_+_26040518 | 2.66 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr11_-_24300905 | 2.65 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr15_+_5124897 | 2.64 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr6_+_50452131 | 2.64 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr9_-_11615763 | 2.62 |
ENSDART00000110245
|
umps
|
uridine monophosphate synthetase |
| chr13_+_35799602 | 2.58 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr19_-_11097075 | 2.57 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr7_-_31346802 | 2.51 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr4_-_12287171 | 2.51 |
ENSDART00000022646
|
cnot4b
|
CCR4-NOT transcription complex, subunit 4b |
| chr5_+_37144384 | 2.46 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr16_+_23976353 | 2.45 |
ENSDART00000058970
|
gtpbp10
|
GTP-binding protein 10 (putative) |
| chr24_+_34183557 | 2.42 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr7_-_41446119 | 2.42 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr10_-_7862983 | 2.42 |
ENSDART00000160606
ENSDART00000112201 |
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr8_-_53165501 | 2.41 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_+_5124690 | 2.40 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr10_-_39362315 | 2.38 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr14_+_15845853 | 2.38 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr19_+_41834452 | 2.37 |
ENSDART00000148732
ENSDART00000149799 |
dlx6a
|
distal-less homeobox 6a |
| chr15_-_33669618 | 2.34 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr7_+_31750808 | 2.30 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr16_-_38383995 | 2.29 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr8_-_22266041 | 2.20 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr6_+_48978653 | 2.20 |
|
|
|
| chr7_-_41532268 | 2.20 |
ENSDART00000016105
ENSDART00000138800 |
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr5_-_24039681 | 2.20 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr14_-_46608307 | 2.19 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr8_-_38284748 | 2.16 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr7_-_31346844 | 2.15 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr8_+_26040405 | 2.13 |
ENSDART00000164207
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr13_+_39151166 | 2.11 |
ENSDART00000113259
|
ENSDARG00000079055
|
ENSDARG00000079055 |
| chr9_+_21548183 | 2.10 |
ENSDART00000059402
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| KN149704v1_-_17031 | 2.10 |
|
|
|
| chr3_-_18655432 | 2.09 |
ENSDART00000034489
|
msrb1a
|
methionine sulfoxide reductase B1a |
| chr16_-_31834774 | 2.04 |
|
|
|
| chr18_+_8959686 | 2.04 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr5_+_12336846 | 2.04 |
ENSDART00000023101
|
pes
|
pescadillo |
| chr5_-_45358634 | 2.04 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr7_+_31750775 | 2.02 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr24_+_39631194 | 2.02 |
ENSDART00000013894
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr5_-_11162837 | 2.01 |
|
|
|
| chr7_+_41532360 | 2.00 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr9_+_27909662 | 1.99 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr14_-_44844086 | 1.98 |
ENSDART00000159988
|
fam193b
|
family with sequence similarity 193, member B |
| chr20_+_2625716 | 1.97 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr24_-_38757042 | 1.96 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr14_+_21779325 | 1.93 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr7_+_29931094 | 1.93 |
ENSDART00000173525
|
wdr76
|
WD repeat domain 76 |
| chr19_+_39689450 | 1.92 |
|
|
|
| chr25_-_8548210 | 1.90 |
ENSDART00000155280
|
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr6_-_22060441 | 1.90 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr5_+_36168475 | 1.89 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr14_-_35932521 | 1.88 |
ENSDART00000158722
|
BX511223.1
|
ENSDARG00000101064 |
| chr19_+_24741351 | 1.85 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr12_-_4596004 | 1.85 |
ENSDART00000160617
|
prr12a
|
proline rich 12a |
| chr13_-_23626319 | 1.84 |
ENSDART00000124152
|
rgs17
|
regulator of G protein signaling 17 |
| chr8_-_38284904 | 1.83 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr9_-_30174641 | 1.82 |
ENSDART00000134157
ENSDART00000089206 |
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr9_+_21548069 | 1.79 |
ENSDART00000147619
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr24_-_21788942 | 1.78 |
ENSDART00000113092
|
abhd10b
|
abhydrolase domain containing 10b |
| chr18_+_27457678 | 1.76 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr5_-_31738565 | 1.76 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_-_13025193 | 1.76 |
ENSDART00000177894
|
BX119963.3
|
ENSDARG00000106390 |
| chr7_+_28782077 | 1.75 |
ENSDART00000134332
|
fam60al
|
family with sequence similarity 60, member A, like |
| chr23_+_38335290 | 1.74 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr13_+_35799851 | 1.74 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr25_-_8548430 | 1.73 |
ENSDART00000155280
|
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr7_-_41446221 | 1.73 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr5_+_69178270 | 1.73 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr9_+_21548044 | 1.73 |
ENSDART00000147619
|
n6amt2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr14_+_15845943 | 1.73 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr20_+_27088328 | 1.73 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr19_-_34529880 | 1.72 |
ENSDART00000158677
ENSDART00000112004 |
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr25_-_7539433 | 1.72 |
ENSDART00000131415
|
arpp19b
|
cAMP-regulated phosphoprotein 19b |
| chr5_-_27394361 | 1.72 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr13_-_4863766 | 1.71 |
ENSDART00000159679
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr21_-_11877525 | 1.71 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr15_+_40228382 | 1.70 |
|
|
|
| chr12_-_21562929 | 1.70 |
ENSDART00000152999
|
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr15_-_33669589 | 1.69 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr19_-_3849836 | 1.68 |
ENSDART00000172415
|
btr18
|
bloodthirsty-related gene family, member 18 |
| chr17_+_23534953 | 1.68 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr19_-_24971920 | 1.68 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr13_-_2296041 | 1.68 |
ENSDART00000126697
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr18_-_20476969 | 1.67 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr7_+_14290727 | 1.67 |
ENSDART00000027331
|
mrpl46
|
mitochondrial ribosomal protein L46 |
| chr1_-_25982625 | 1.65 |
ENSDART00000152492
|
trmo
|
tRNA methyltransferase O |
| chr20_-_46079578 | 1.64 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr5_-_12592887 | 1.64 |
|
|
|
| chr5_-_47609309 | 1.63 |
|
|
|
| chr1_-_53021646 | 1.63 |
|
|
|
| chr17_-_7661725 | 1.63 |
ENSDART00000143870
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr22_-_10562118 | 1.62 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr4_-_17654047 | 1.62 |
|
|
|
| chr20_+_25726978 | 1.62 |
ENSDART00000164264
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr21_-_44086464 | 1.61 |
ENSDART00000130833
|
FO704810.1
|
ENSDARG00000087492 |
| chr22_-_36721626 | 1.61 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr3_+_31530419 | 1.61 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr20_+_38354928 | 1.60 |
ENSDART00000149160
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr2_+_19588034 | 1.60 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr13_-_11888864 | 1.59 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr19_+_43523515 | 1.58 |
ENSDART00000023156
ENSDART00000039129 |
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr5_-_18561185 | 1.58 |
|
|
|
| chr12_+_18776712 | 1.55 |
|
|
|
| chr4_-_1496779 | 1.54 |
ENSDART00000166360
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_54551887 | 1.53 |
ENSDART00000169663
|
CT573860.1
|
ENSDARG00000098327 |
| chr20_-_3292418 | 1.53 |
ENSDART00000099315
|
cebpz
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr21_-_22085596 | 1.53 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr10_-_7862767 | 1.52 |
ENSDART00000059021
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr24_+_39630741 | 1.51 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr11_-_24300849 | 1.51 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr15_+_40228295 | 1.51 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr11_+_3939876 | 1.49 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr1_-_42915971 | 1.49 |
|
|
|
| chr15_+_5124820 | 1.48 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr1_+_5437488 | 1.48 |
ENSDART00000092324
|
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr24_-_38757086 | 1.48 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr5_+_68036497 | 1.48 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr5_-_47609402 | 1.47 |
|
|
|
| chr8_-_38284959 | 1.47 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr10_-_15382145 | 1.47 |
ENSDART00000133919
|
pum3
|
pumilio RNA-binding family member 3 |
| chr16_-_42015364 | 1.47 |
ENSDART00000136742
|
BX649388.1
|
ENSDARG00000091933 |
| chr4_+_17653739 | 1.46 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr13_+_6060281 | 1.46 |
ENSDART00000161062
ENSDART00000109665 |
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr19_-_2084484 | 1.45 |
|
|
|
| chr2_+_10192670 | 1.44 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr13_+_39151116 | 1.44 |
ENSDART00000113259
|
ENSDARG00000079055
|
ENSDARG00000079055 |
| chr22_-_36721592 | 1.43 |
ENSDART00000130171
|
ncl
|
nucleolin |
| chr5_+_12336706 | 1.43 |
ENSDART00000023101
|
pes
|
pescadillo |
| chr4_-_7721419 | 1.43 |
|
|
|
| chr4_-_14193396 | 1.43 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr3_-_35734943 | 1.42 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr19_+_42758938 | 1.42 |
ENSDART00000077059
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr25_+_21001338 | 1.41 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr1_-_19071020 | 1.41 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr24_+_36430199 | 1.40 |
|
|
|
| chr3_-_56803756 | 1.40 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr17_-_29196678 | 1.40 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr18_+_6339577 | 1.40 |
ENSDART00000136333
|
wash1
|
WAS protein family homolog 1 |
| chr20_-_53123015 | 1.39 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_-_5153412 | 1.39 |
ENSDART00000160037
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr23_+_37124543 | 1.39 |
ENSDART00000155706
|
CR762407.2
|
ENSDARG00000097111 |
| chr8_-_22267016 | 1.39 |
ENSDART00000140978
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr25_+_18997440 | 1.39 |
ENSDART00000155569
|
si:dkeyp-19h3.8
|
si:dkeyp-19h3.8 |
| chr23_-_21607742 | 1.39 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr8_-_22266750 | 1.39 |
ENSDART00000100042
ENSDART00000138966 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr20_-_22299344 | 1.38 |
ENSDART00000063578
|
srd5a3
|
steroid 5 alpha-reductase 3 |
| chr6_-_22059932 | 1.38 |
ENSDART00000162148
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr8_+_31019730 | 1.38 |
|
|
|
| chr3_-_5523147 | 1.38 |
ENSDART00000159308
|
trim35-7
|
tripartite motif containing 35-7 |
| chr6_+_58528496 | 1.38 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr8_+_29885318 | 1.37 |
ENSDART00000154309
|
CR376747.1
|
ENSDARG00000097806 |
| chr25_+_21001016 | 1.37 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr19_-_2222108 | 1.37 |
ENSDART00000127578
|
twistnb
|
TWIST neighbor |
| chr16_-_13723778 | 1.36 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr13_+_8560739 | 1.36 |
ENSDART00000075054
|
thada
|
thyroid adenoma associated |
| chr24_-_6647047 | 1.36 |
|
|
|
| chr3_+_7877162 | 1.36 |
ENSDART00000057434
ENSDART00000170291 |
hook2
|
hook microtubule-tethering protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 1.9 | 5.8 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 1.9 | 7.5 | GO:1990379 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 1.2 | 6.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.1 | 4.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 1.1 | 4.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.9 | 5.6 | GO:0045002 | DNA recombinase assembly(GO:0000730) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.9 | 4.4 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.8 | 3.2 | GO:0036049 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.8 | 3.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.8 | 8.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.7 | 2.9 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.7 | 2.9 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.7 | 5.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.6 | 1.9 | GO:0050857 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.6 | 5.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.6 | 1.8 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.5 | 3.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.5 | 3.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 3.3 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.5 | 2.7 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.4 | 4.8 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.4 | 5.6 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.4 | 2.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 1.3 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.4 | 0.8 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.4 | 4.6 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.4 | 1.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.4 | 0.4 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.4 | 4.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 5.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 1.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 1.4 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.3 | 1.0 | GO:0046887 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.3 | 4.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 1.6 | GO:0071346 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.3 | 1.9 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.3 | 0.9 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.3 | 1.1 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.3 | 1.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.3 | 0.8 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.3 | 4.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.3 | 0.8 | GO:0071028 | U4 snRNA 3'-end processing(GO:0034475) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 6.5 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.2 | 3.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.2 | 1.8 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.2 | 11.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.2 | 0.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 10.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 3.5 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.2 | 1.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 0.6 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 1.4 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.0 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.2 | 1.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.2 | 6.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 3.7 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.2 | 6.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.2 | 0.7 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.2 | 1.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 6.3 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.2 | 2.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 2.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 1.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 2.4 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.2 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.9 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.6 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 2.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.7 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.1 | 0.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 1.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.8 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.4 | GO:0045943 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.4 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) hyaluronan biosynthetic process(GO:0030213) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 2.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 3.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.9 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.4 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.1 | 3.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.7 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 1.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.5 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0070661 | mononuclear cell proliferation(GO:0032943) T cell proliferation(GO:0042098) lymphocyte proliferation(GO:0046651) leukocyte proliferation(GO:0070661) |
| 0.0 | 0.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 2.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 3.8 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.6 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 1.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.1 | GO:0048477 | female gamete generation(GO:0007292) oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0006555 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.7 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.2 | GO:0070646 | protein modification by small protein removal(GO:0070646) |
| 0.0 | 0.2 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.2 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.6 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 10.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.9 | 3.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.7 | 2.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.7 | 4.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.7 | 3.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.6 | 3.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.6 | 3.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.6 | 5.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 4.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 1.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 3.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 2.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.4 | 2.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 5.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 0.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 6.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 2.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 2.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 2.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 2.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 7.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 3.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 31.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 2.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 2.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic recycling endosome(GO:0098837) postsynaptic endosome(GO:0098845) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.3 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 9.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 1.5 | 4.6 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 1.4 | 5.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.1 | 3.2 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.9 | 4.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.8 | 4.9 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.8 | 3.2 | GO:0036055 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.8 | 4.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.8 | 4.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.7 | 2.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.7 | 3.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.6 | 1.9 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.6 | 1.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.5 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 9.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.5 | 11.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.5 | 1.4 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.5 | 4.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.5 | 1.4 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.4 | 3.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 5.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.4 | 1.3 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.4 | 1.2 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.4 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 1.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 6.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 0.9 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.3 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 3.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 0.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 3.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 3.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 1.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 4.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 2.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 1.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 1.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 6.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 7.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 0.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 1.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 4.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 1.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 1.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.7 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 16.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.2 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 4.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 4.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 13.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 5.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 3.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 2.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 7.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 5.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.7 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.1 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 4.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.5 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.0 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 9.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 2.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 6.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 1.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 1.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 3.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 5.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 4.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 1.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 10.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 1.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 2.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |