Project
DANIO-CODE
Navigation
Downloads
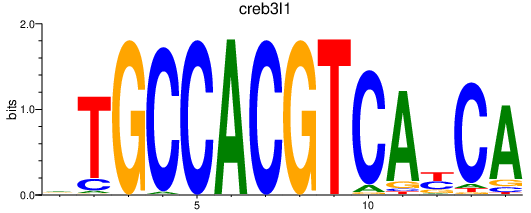
Results for creb3l1
Z-value: 0.66
Transcription factors associated with creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l1
|
ENSDARG00000015793 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l1 | dr10_dc_chr7_+_38626646_38626717 | -0.02 | 9.3e-01 | Click! |
Activity profile of creb3l1 motif
Sorted Z-values of creb3l1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_9934584 | 1.37 |
ENSDART00000169522
ENSDART00000160156 |
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr18_-_34573597 | 1.30 |
ENSDART00000021880
|
ssr3
|
signal sequence receptor, gamma |
| chr17_+_9934845 | 1.21 |
ENSDART00000168055
|
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr5_-_29916352 | 1.16 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr10_+_15644868 | 1.13 |
ENSDART00000139259
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr18_+_18598885 | 1.00 |
|
|
|
| chr21_-_43641419 | 0.96 |
ENSDART00000151486
ENSDART00000151115 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr11_-_18818010 | 0.93 |
|
|
|
| chr20_-_28505341 | 0.73 |
ENSDART00000055932
|
pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr7_+_17848688 | 0.63 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr21_+_10609580 | 0.60 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr13_+_7924797 | 0.58 |
|
|
|
| chr20_+_22900165 | 0.58 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr1_-_54453712 | 0.51 |
ENSDART00000074083
|
rab1aa
|
RAB1A, member RAS oncogene family a |
| chr3_+_32731002 | 0.45 |
ENSDART00000137599
|
zgc:162613
|
zgc:162613 |
| chr14_-_46477165 | 0.44 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr9_-_20743808 | 0.43 |
|
|
|
| chr25_-_8436134 | 0.42 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr18_-_34573536 | 0.41 |
ENSDART00000137101
|
ssr3
|
signal sequence receptor, gamma |
| chr20_-_1170970 | 0.40 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr6_-_21756764 | 0.34 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr3_+_34690515 | 0.32 |
|
|
|
| chr14_-_46476712 | 0.32 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr5_-_1432791 | 0.29 |
ENSDART00000128274
|
golga2
|
golgin A2 |
| chr6_-_9000728 | 0.29 |
ENSDART00000160397
|
iqcb1
|
IQ motif containing B1 |
| chr8_+_28377022 | 0.28 |
ENSDART00000158788
|
klhl12
|
kelch-like family member 12 |
| chr18_+_7595039 | 0.27 |
ENSDART00000062143
|
zgc:77650
|
zgc:77650 |
| chr12_-_17357133 | 0.27 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr3_+_34690574 | 0.26 |
|
|
|
| chr8_+_29885318 | 0.26 |
ENSDART00000154309
|
CR376747.1
|
ENSDARG00000097806 |
| chr18_-_15300493 | 0.25 |
ENSDART00000048206
|
tmem263
|
transmembrane protein 263 |
| chr14_-_46476881 | 0.25 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr19_+_21124187 | 0.25 |
ENSDART00000129730
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr20_+_22900302 | 0.24 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr18_-_7522063 | 0.23 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr9_+_20743306 | 0.23 |
ENSDART00000147435
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr18_-_7072418 | 0.23 |
ENSDART00000148485
|
calub
|
calumenin b |
| chr23_-_31339986 | 0.23 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr21_+_10609341 | 0.22 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr17_+_30431395 | 0.22 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr25_-_2908684 | 0.22 |
ENSDART00000073618
|
scamp2
|
secretory carrier membrane protein 2 |
| chr15_-_29454583 | 0.22 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr15_-_18179431 | 0.21 |
ENSDART00000047902
|
arcn1b
|
archain 1b |
| chr6_-_21756853 | 0.20 |
ENSDART00000169035
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr7_-_29300402 | 0.20 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr19_+_33551956 | 0.20 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr16_+_5712858 | 0.19 |
ENSDART00000140226
|
CYTH2
|
cytohesin 2 |
| chr19_+_7630609 | 0.19 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr7_-_58523645 | 0.18 |
|
|
|
| chr21_-_32403403 | 0.18 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr7_-_29300448 | 0.17 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr15_+_23849554 | 0.17 |
ENSDART00000138375
|
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr8_-_37429347 | 0.16 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr20_+_38717806 | 0.16 |
ENSDART00000045850
|
si:ch211-243j20.2
|
si:ch211-243j20.2 |
| chr7_+_51550533 | 0.16 |
ENSDART00000174097
|
BX957362.1
|
ENSDARG00000105682 |
| chr21_-_11870303 | 0.15 |
ENSDART00000123335
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr2_+_24152953 | 0.15 |
ENSDART00000131030
|
gorasp1a
|
golgi reassembly stacking protein 1a |
| chr14_+_8288174 | 0.14 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr4_+_25662491 | 0.14 |
ENSDART00000100693
|
acot16
|
acyl-CoA thioesterase 16 |
| chr24_+_28441089 | 0.13 |
|
|
|
| chr7_+_13238684 | 0.13 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr4_+_11465367 | 0.13 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr6_+_202117 | 0.12 |
|
|
|
| chr13_-_9543393 | 0.11 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr23_+_18565721 | 0.10 |
|
|
|
| chr14_-_26113593 | 0.09 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr19_+_7630847 | 0.09 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr16_-_20044830 | 0.09 |
|
|
|
| chr10_-_3427119 | 0.09 |
ENSDART00000037183
|
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr11_-_18393802 | 0.08 |
ENSDART00000125453
|
dido1
|
death inducer-obliterator 1 |
| chr1_-_49004615 | 0.08 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr23_-_42775849 | 0.06 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr10_+_31701855 | 0.06 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr1_-_19001718 | 0.06 |
ENSDART00000143303
|
BX323590.2
|
ENSDARG00000095388 |
| chr20_-_3238990 | 0.06 |
|
|
|
| chr2_-_23516930 | 0.06 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr14_-_46608307 | 0.06 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr22_-_31110564 | 0.04 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr3_-_22699157 | 0.04 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr19_+_42763897 | 0.02 |
|
|
|
| chr6_+_3703684 | 0.01 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr22_+_31110716 | 0.01 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr2_-_23516991 | 0.01 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr18_-_15300414 | 0.01 |
ENSDART00000048206
|
tmem263
|
transmembrane protein 263 |
| chr19_+_7630777 | 0.01 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr5_+_44208406 | 0.00 |
|
|
|
| chr4_-_76594399 | 0.00 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.2 | 1.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.3 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.2 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.4 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.3 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0048200 | vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.0 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.8 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |