Project
DANIO-CODE
Navigation
Downloads
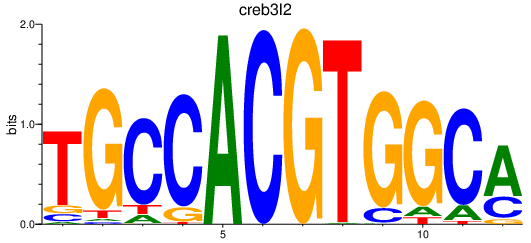
Results for creb3l2
Z-value: 0.67
Transcription factors associated with creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
creb3l2
|
ENSDARG00000063563 | cAMP responsive element binding protein 3-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| creb3l2 | dr10_dc_chr4_-_4743320_4743409 | -0.52 | 4.0e-02 | Click! |
Activity profile of creb3l2 motif
Sorted Z-values of creb3l2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of creb3l2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_59972297 | 0.94 |
|
|
|
| chr2_-_6153363 | 0.65 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr20_+_32503118 | 0.63 |
ENSDART00000018640
|
snx3
|
sorting nexin 3 |
| chr7_+_15479700 | 0.62 |
|
|
|
| chr17_+_28689850 | 0.60 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr3_+_42217236 | 0.60 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr14_+_47040687 | 0.59 |
ENSDART00000113469
|
nocta
|
nocturnin a |
| chr20_+_51248443 | 0.59 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr18_-_37271104 | 0.59 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr10_-_44441390 | 0.58 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr17_+_25815708 | 0.57 |
ENSDART00000164160
|
acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr25_-_7490206 | 0.56 |
ENSDART00000163018
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr2_+_51265445 | 0.53 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr7_-_27787236 | 0.51 |
ENSDART00000173842
|
RIC3
|
si:ch211-235p24.2 |
| chr4_+_76572863 | 0.50 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr25_+_35782970 | 0.45 |
ENSDART00000125440
|
hist1h4l
|
histone 1, H4, like |
| chr8_+_8660457 | 0.45 |
ENSDART00000137780
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr5_-_54091790 | 0.44 |
ENSDART00000150070
|
ccnb1
|
cyclin B1 |
| chr25_-_1243081 | 0.43 |
ENSDART00000156062
|
calml4b
|
calmodulin-like 4b |
| chr11_+_25058986 | 0.42 |
ENSDART00000122249
|
top1l
|
topoisomerase (DNA) I, like |
| chr23_+_38335290 | 0.40 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr1_-_55072271 | 0.39 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr11_+_25058714 | 0.39 |
ENSDART00000065949
|
top1l
|
topoisomerase (DNA) I, like |
| chr25_-_34616837 | 0.39 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr10_-_1185831 | 0.38 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr18_-_37271027 | 0.38 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr7_-_72161793 | 0.37 |
ENSDART00000162933
|
mrpl21
|
mitochondrial ribosomal protein L21 |
| chr20_+_29306863 | 0.37 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_-_21627098 | 0.37 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr4_-_16893974 | 0.36 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr13_+_36831946 | 0.33 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr7_+_38444768 | 0.33 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr19_+_7254917 | 0.33 |
ENSDART00000123934
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr20_+_29307039 | 0.32 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_-_36499541 | 0.32 |
|
|
|
| chr21_-_5774127 | 0.32 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr17_-_28780394 | 0.32 |
ENSDART00000167897
ENSDART00000134735 |
scfd1
|
sec1 family domain containing 1 |
| chr5_-_54091732 | 0.31 |
ENSDART00000150070
|
ccnb1
|
cyclin B1 |
| chr2_-_6153411 | 0.30 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr3_-_15913190 | 0.30 |
|
|
|
| chr20_+_15666406 | 0.30 |
ENSDART00000063917
|
si:dkey-86e18.1
|
si:dkey-86e18.1 |
| chr20_-_46079578 | 0.29 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr1_-_53021646 | 0.29 |
|
|
|
| chr13_-_25418278 | 0.28 |
ENSDART00000077655
ENSDART00000168099 ENSDART00000135788 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr20_+_29306677 | 0.28 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr16_-_31664756 | 0.27 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr17_-_44326992 | 0.27 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr2_-_39049391 | 0.27 |
ENSDART00000044331
|
copb2
|
coatomer protein complex, subunit beta 2 |
| chr20_+_29306945 | 0.27 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_+_34563757 | 0.27 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr7_+_15059782 | 0.27 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr20_-_46079529 | 0.26 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr3_+_43361682 | 0.26 |
ENSDART00000168784
|
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr2_+_54267946 | 0.25 |
ENSDART00000015535
|
ctnnbl1
|
catenin, beta like 1 |
| chr3_+_42217187 | 0.24 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr22_+_31110716 | 0.24 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr20_-_13244237 | 0.24 |
ENSDART00000124470
|
ints7
|
integrator complex subunit 7 |
| chr14_+_2820269 | 0.24 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr25_-_7546532 | 0.23 |
ENSDART00000131583
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr21_+_45622590 | 0.22 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr20_-_7079412 | 0.22 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr3_+_22204475 | 0.21 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr16_-_20488542 | 0.21 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr8_+_8660574 | 0.21 |
ENSDART00000137780
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr13_-_25452733 | 0.21 |
|
|
|
| chr3_+_43361636 | 0.21 |
ENSDART00000161277
|
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr15_+_666833 | 0.20 |
ENSDART00000157207
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr2_-_55565099 | 0.20 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr15_+_32561282 | 0.20 |
ENSDART00000162663
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr2_-_45118469 | 0.20 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr5_+_1737449 | 0.19 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr7_-_69114957 | 0.19 |
|
|
|
| chr15_+_23272957 | 0.19 |
ENSDART00000006085
|
cbl
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr6_+_251026 | 0.19 |
ENSDART00000042970
|
atf4a
|
activating transcription factor 4a |
| chr13_-_29571309 | 0.19 |
ENSDART00000139773
|
CT025909.1
|
ENSDARG00000093614 |
| chr14_+_47040931 | 0.19 |
ENSDART00000113469
|
nocta
|
nocturnin a |
| chr5_+_59394222 | 0.19 |
|
|
|
| chr20_+_29307283 | 0.18 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_-_21779536 | 0.18 |
|
|
|
| chr13_+_36832145 | 0.17 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr14_-_35513309 | 0.17 |
ENSDART00000173343
|
pdgfc
|
platelet derived growth factor c |
| chr15_+_1569674 | 0.17 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr1_-_55072064 | 0.17 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr24_-_33475705 | 0.17 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr13_+_36832090 | 0.17 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr25_-_20633069 | 0.17 |
ENSDART00000172170
|
si:ch211-127m7.2
|
si:ch211-127m7.2 |
| chr16_+_20488737 | 0.16 |
ENSDART00000059619
|
fkbp14
|
FK506 binding protein 14 |
| chr12_-_9094888 | 0.16 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr20_+_29307142 | 0.16 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| KN150699v1_+_15349 | 0.16 |
ENSDART00000167656
|
creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr8_+_23192085 | 0.16 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr8_-_17736487 | 0.15 |
ENSDART00000063592
ENSDART00000175064 |
prkcz
|
protein kinase C, zeta |
| chr5_+_68505700 | 0.14 |
ENSDART00000136656
|
selm
|
selenoprotein M |
| chr20_-_7079708 | 0.13 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr2_-_10602948 | 0.13 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr24_+_23599223 | 0.12 |
|
|
|
| chr19_-_25173832 | 0.12 |
ENSDART00000161016
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr7_+_69664384 | 0.12 |
ENSDART00000058763
|
sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr3_+_42217445 | 0.12 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr21_-_19883010 | 0.12 |
ENSDART00000137307
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr11_-_7146819 | 0.11 |
ENSDART00000172879
|
smim7
|
small integral membrane protein 7 |
| chr3_-_18234622 | 0.11 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr21_+_45622095 | 0.10 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr21_-_37404533 | 0.10 |
ENSDART00000129439
|
BX649250.1
|
ENSDARG00000091187 |
| chr23_+_38335341 | 0.09 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr4_+_344355 | 0.09 |
ENSDART00000142225
ENSDART00000163436 |
tmem181
|
transmembrane protein 181 |
| chr8_-_17736169 | 0.09 |
ENSDART00000063592
ENSDART00000175064 |
prkcz
|
protein kinase C, zeta |
| chr11_-_7146782 | 0.09 |
ENSDART00000172823
|
smim7
|
small integral membrane protein 7 |
| chr1_+_25750825 | 0.08 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr21_-_3462128 | 0.08 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr20_-_7079663 | 0.08 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr14_+_21779325 | 0.08 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr5_-_28368834 | 0.08 |
|
|
|
| chr3_+_39426379 | 0.07 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr17_+_28690237 | 0.07 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr4_-_23028510 | 0.07 |
ENSDART00000040033
ENSDART00000160375 |
nup107
|
nucleoporin 107 |
| chr8_-_8659980 | 0.07 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr20_-_20412830 | 0.07 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr7_+_15479520 | 0.07 |
|
|
|
| chr13_-_40189984 | 0.06 |
|
|
|
| chr22_-_800449 | 0.06 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr17_+_44327269 | 0.06 |
ENSDART00000045882
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr19_+_47846675 | 0.06 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr1_-_25750237 | 0.06 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr10_-_44441481 | 0.06 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr12_+_33793495 | 0.06 |
ENSDART00000152988
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr25_-_36512943 | 0.06 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr17_-_28794581 | 0.06 |
ENSDART00000132811
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr22_-_4715399 | 0.04 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr3_+_22204569 | 0.03 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr15_-_43954665 | 0.03 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr3_+_22724101 | 0.03 |
ENSDART00000167127
|
BX322574.1
|
ENSDARG00000098353 |
| chr10_-_44441197 | 0.03 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr20_-_13244278 | 0.02 |
ENSDART00000124470
|
ints7
|
integrator complex subunit 7 |
| chr5_+_56658275 | 0.02 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr25_+_34357376 | 0.02 |
ENSDART00000149782
|
CHST6
|
carbohydrate sulfotransferase 6 |
| chr10_-_45535333 | 0.02 |
|
|
|
| chr21_-_45621982 | 0.02 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr5_-_62963536 | 0.01 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr2_-_11720585 | 0.01 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr2_+_54268010 | 0.01 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr8_+_46378250 | 0.01 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr8_-_46037132 | 0.00 |
ENSDART00000135138
|
si:ch211-119d14.3
|
si:ch211-119d14.3 |
| chr17_-_28794619 | 0.00 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.0 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.1 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.4 | GO:0036049 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.1 | 0.6 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.3 | GO:0070863 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.0 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.7 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.0 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0035620 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |