Project
DANIO-CODE
Navigation
Downloads
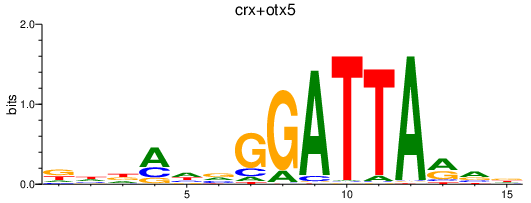
Results for crx+otx5
Z-value: 1.48
Transcription factors associated with crx+otx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
crx
|
ENSDARG00000011989 | cone-rod homeobox |
|
otx5
|
ENSDARG00000043483 | orthodenticle homolog 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx5 | dr10_dc_chr15_+_47316316_47316434 | -0.33 | 2.2e-01 | Click! |
| crx | dr10_dc_chr5_+_36332564_36332612 | -0.29 | 2.7e-01 | Click! |
Activity profile of crx+otx5 motif
Sorted Z-values of crx+otx5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of crx+otx5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_4264663 | 6.81 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| chr20_+_9141086 | 4.12 |
ENSDART00000064144
ENSDART00000144396 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr14_+_44703094 | 3.39 |
|
|
|
| chr15_+_44177774 | 3.05 |
|
|
|
| chr9_+_18821640 | 2.98 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr15_+_44177693 | 2.97 |
|
|
|
| chr17_+_16038358 | 2.91 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr12_-_13866903 | 2.91 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr3_-_26052785 | 2.69 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr22_+_25113293 | 2.67 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr23_+_39961676 | 2.51 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| chr11_-_2437396 | 2.49 |
|
|
|
| chr15_+_40232397 | 2.34 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr12_-_13866811 | 2.29 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr8_-_3354396 | 2.24 |
ENSDART00000169430
ENSDART00000167187 ENSDART00000170478 |
FUT9 (1 of many)
fut9b
|
fucosyltransferase 9 fucosyltransferase 9b |
| chr1_+_39315964 | 2.16 |
ENSDART00000011727
|
ccdc149b
|
coiled-coil domain containing 149b |
| chr25_-_36512943 | 2.10 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr17_+_16038103 | 1.98 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr4_-_3184182 | 1.95 |
|
|
|
| chr3_-_26052601 | 1.95 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_+_38497816 | 1.95 |
|
|
|
| chr24_+_36429525 | 1.92 |
ENSDART00000062722
|
pus3
|
pseudouridylate synthase 3 |
| chr19_-_2148053 | 1.90 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr5_-_68408107 | 1.88 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr3_+_16692138 | 1.84 |
ENSDART00000023985
|
zgc:153952
|
zgc:153952 |
| chr3_+_16692090 | 1.82 |
ENSDART00000023985
|
zgc:153952
|
zgc:153952 |
| chr3_+_17783319 | 1.81 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr8_+_25940758 | 1.74 |
ENSDART00000140626
|
ENSDARG00000061023
|
ENSDARG00000061023 |
| chr24_+_10758290 | 1.73 |
ENSDART00000106272
|
si:dkey-37o8.1
|
si:dkey-37o8.1 |
| chr17_+_50622734 | 1.70 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr16_+_25370991 | 1.68 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr20_-_15205248 | 1.66 |
ENSDART00000034011
|
fmo5
|
flavin containing monooxygenase 5 |
| chr24_+_39237314 | 1.64 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr13_+_40689102 | 1.60 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr10_-_1933761 | 1.58 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr12_-_4264610 | 1.54 |
ENSDART00000152377
|
ca15b
|
carbonic anhydrase XVb |
| chr15_+_23721725 | 1.53 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr24_-_28227116 | 1.52 |
ENSDART00000148618
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr20_-_34851706 | 1.48 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr10_-_25725495 | 1.44 |
ENSDART00000177220
|
AL837524.2
|
ENSDARG00000108650 |
| chr24_+_16861058 | 1.42 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr19_-_9584480 | 1.42 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr25_-_31352866 | 1.41 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr20_+_35156812 | 1.40 |
|
|
|
| chr8_+_36527653 | 1.40 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr8_+_2484943 | 1.39 |
ENSDART00000101137
|
gle1
|
GLE1 RNA export mediator homolog (yeast) |
| chr23_-_24328931 | 1.38 |
|
|
|
| chr14_-_16118942 | 1.37 |
|
|
|
| chr24_+_16861121 | 1.36 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr24_-_21224397 | 1.36 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr3_+_25776714 | 1.35 |
ENSDART00000170324
|
tom1
|
target of myb1 membrane trafficking protein |
| chr20_+_6543305 | 1.34 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr11_-_2437361 | 1.31 |
|
|
|
| chr14_+_1106141 | 1.30 |
ENSDART00000106708
|
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr19_+_10742944 | 1.27 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr16_+_40093596 | 1.27 |
ENSDART00000132457
|
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr2_-_4191540 | 1.26 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr9_+_42656327 | 1.26 |
|
|
|
| chr7_-_41601001 | 1.25 |
ENSDART00000174258
|
zgc:92818
|
zgc:92818 |
| chr24_+_440200 | 1.22 |
|
|
|
| chr11_-_11353309 | 1.22 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr13_-_35401769 | 1.22 |
ENSDART00000016793
|
memo1
|
mediator of cell motility 1 |
| chr24_-_21224371 | 1.20 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr11_-_6858626 | 1.19 |
ENSDART00000168372
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_-_45244910 | 1.19 |
|
|
|
| chr10_+_20435169 | 1.17 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr8_-_3354350 | 1.16 |
ENSDART00000169430
ENSDART00000167187 |
FUT9 (1 of many)
|
fucosyltransferase 9 |
| chr10_-_25059072 | 1.14 |
ENSDART00000040733
|
cd209
|
CD209 molecule |
| chr24_+_36430199 | 1.13 |
|
|
|
| chr5_-_23525132 | 1.13 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr22_+_26270259 | 1.13 |
ENSDART00000060898
|
mrps28
|
mitochondrial ribosomal protein S28 |
| chr5_+_29301998 | 1.12 |
ENSDART00000175940
|
BX284646.1
|
ENSDARG00000106174 |
| chr19_+_16159980 | 1.11 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr25_+_181958 | 1.09 |
ENSDART00000155412
|
RPS17
|
ribosomal protein S17 |
| chr21_+_3624059 | 1.07 |
ENSDART00000111699
|
tor1
|
torsin family 1 |
| chr8_-_50151276 | 1.07 |
|
|
|
| chr17_+_25407766 | 1.06 |
ENSDART00000121848
|
srrm1
|
serine/arginine repetitive matrix 1 |
| chr24_+_35296732 | 1.06 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr19_-_25842574 | 1.03 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr5_-_22090338 | 1.02 |
|
|
|
| chr3_+_48811909 | 0.97 |
ENSDART00000136051
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr24_+_35297193 | 0.97 |
ENSDART00000075142
ENSDART00000158801 |
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr3_+_23963926 | 0.96 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr16_+_25370955 | 0.94 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr23_+_19271879 | 0.93 |
ENSDART00000047015
|
ccdc115
|
coiled-coil domain containing 115 |
| chr9_-_12603178 | 0.90 |
|
|
|
| chr8_-_28209155 | 0.90 |
ENSDART00000154912
|
BX664742.2
|
ENSDARG00000095929 |
| chr16_-_40093241 | 0.89 |
ENSDART00000145278
|
si:dkey-29b11.3
|
si:dkey-29b11.3 |
| chr17_+_26736703 | 0.89 |
|
|
|
| chr20_-_43889302 | 0.89 |
ENSDART00000004601
|
laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr25_-_34235582 | 0.88 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr24_-_20782338 | 0.88 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr8_+_2597651 | 0.88 |
ENSDART00000165943
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr10_-_1933874 | 0.88 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr15_+_34106908 | 0.87 |
|
|
|
| chr3_-_20807407 | 0.87 |
ENSDART00000164601
|
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr3_+_48811632 | 0.86 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr11_+_6872272 | 0.86 |
ENSDART00000075998
|
klhl26
|
kelch-like family member 26 |
| chr1_+_35224410 | 0.85 |
ENSDART00000085021
ENSDART00000140144 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr17_-_8435280 | 0.84 |
ENSDART00000127022
|
ENSDARG00000090072
|
ENSDARG00000090072 |
| chr11_+_31417477 | 0.84 |
ENSDART00000139900
|
diaph3
|
diaphanous-related formin 3 |
| chr15_+_24012650 | 0.83 |
ENSDART00000153951
|
myo18ab
|
myosin XVIIIAb |
| chr9_-_12603215 | 0.82 |
|
|
|
| chr6_+_40835073 | 0.81 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr12_+_11314303 | 0.80 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr5_-_3255080 | 0.80 |
ENSDART00000152108
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr25_-_34235633 | 0.79 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr4_-_71945633 | 0.78 |
ENSDART00000170170
|
znf1015
|
zinc finger protein 1015 |
| chr2_+_55065658 | 0.77 |
|
|
|
| chr7_+_20829933 | 0.76 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr3_+_48811662 | 0.76 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr22_+_1769284 | 0.76 |
ENSDART00000164948
|
znf1169
|
zinc finger protein 1169 |
| chr13_+_39056158 | 0.74 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr23_-_36350571 | 0.74 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr10_+_6011791 | 0.73 |
ENSDART00000159216
|
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr8_-_41271389 | 0.72 |
ENSDART00000170332
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr18_+_38210277 | 0.72 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr2_+_55112111 | 0.71 |
ENSDART00000163834
|
rnf126
|
ring finger protein 126 |
| chr20_+_48631844 | 0.71 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr20_+_23843178 | 0.70 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr21_-_39616469 | 0.69 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr17_-_25284780 | 0.67 |
ENSDART00000167992
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr25_-_27279272 | 0.67 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr6_-_33940201 | 0.65 |
ENSDART00000132762
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr25_-_36513113 | 0.65 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr19_-_2147899 | 0.63 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr20_+_29685379 | 0.63 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr9_-_7110860 | 0.61 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr3_-_26927151 | 0.61 |
ENSDART00000153542
|
CR954963.1
|
ENSDARG00000097153 |
| chr11_-_18073851 | 0.59 |
|
|
|
| chr2_-_54218077 | 0.59 |
|
|
|
| chr19_-_3947422 | 0.59 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr5_+_34381412 | 0.58 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr11_+_3479741 | 0.56 |
ENSDART00000171621
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr6_-_33940469 | 0.55 |
ENSDART00000137268
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr22_-_26269585 | 0.54 |
ENSDART00000043774
|
sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr18_-_17542397 | 0.53 |
ENSDART00000061000
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr24_+_35296642 | 0.52 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr9_+_3547661 | 0.52 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr7_+_28753102 | 0.52 |
ENSDART00000076425
|
cog4
|
component of oligomeric golgi complex 4 |
| chr15_+_1018191 | 0.51 |
|
|
|
| chr15_-_30975156 | 0.51 |
|
|
|
| chr20_-_18836698 | 0.51 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr20_+_34768464 | 0.51 |
ENSDART00000152836
|
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr14_-_40941231 | 0.50 |
ENSDART00000138917
|
arl13a
|
ADP-ribosylation factor-like 13A |
| chr3_+_23964015 | 0.50 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr11_-_6858313 | 0.50 |
ENSDART00000037824
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_+_36527535 | 0.48 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr15_+_46800117 | 0.48 |
|
|
|
| chr25_+_35771748 | 0.42 |
|
|
|
| chr21_-_13688502 | 0.42 |
ENSDART00000024720
|
si:ch211-282j22.3
|
si:ch211-282j22.3 |
| chr23_-_31340025 | 0.42 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr21_-_13599764 | 0.41 |
|
|
|
| chr16_+_53391438 | 0.38 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr4_+_135858 | 0.37 |
ENSDART00000171845
|
gpr19
|
G protein-coupled receptor 19 |
| chr17_-_49910160 | 0.36 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr16_+_24046996 | 0.32 |
|
|
|
| chr20_+_23843436 | 0.32 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| KN149789v1_-_7443 | 0.31 |
|
|
|
| chr6_-_17699766 | 0.30 |
ENSDART00000170597
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr12_-_33358480 | 0.30 |
ENSDART00000105562
|
zgc:91940
|
zgc:91940 |
| chr13_+_35402382 | 0.30 |
ENSDART00000163368
|
wdr27
|
WD repeat domain 27 |
| chr3_-_26013660 | 0.30 |
|
|
|
| chr3_-_23442774 | 0.29 |
|
|
|
| chr20_-_15205171 | 0.29 |
ENSDART00000034011
|
fmo5
|
flavin containing monooxygenase 5 |
| chr18_+_2902170 | 0.29 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr3_+_19515578 | 0.27 |
ENSDART00000007857
|
mettl2a
|
methyltransferase like 2A |
| chr7_-_66641207 | 0.26 |
|
|
|
| chr7_-_66641971 | 0.25 |
|
|
|
| chr6_-_39767422 | 0.25 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr17_+_35149038 | 0.25 |
ENSDART00000163557
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr3_+_45375580 | 0.23 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr16_-_35462083 | 0.23 |
ENSDART00000170048
|
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_+_21592861 | 0.21 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr21_-_3619164 | 0.20 |
ENSDART00000133980
|
scamp1
|
secretory carrier membrane protein 1 |
| chr1_-_37452112 | 0.19 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr16_+_14311271 | 0.19 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr5_-_3254995 | 0.19 |
ENSDART00000152108
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr5_+_60972907 | 0.15 |
ENSDART00000134387
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr8_-_43568078 | 0.14 |
ENSDART00000051139
ENSDART00000160289 |
ncor2
|
nuclear receptor corepressor 2 |
| chr3_-_23620163 | 0.12 |
|
|
|
| chr9_+_19707706 | 0.12 |
ENSDART00000157395
ENSDART00000102214 |
ndufv3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3 |
| chr16_+_34157948 | 0.11 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr7_-_18916067 | 0.11 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr16_-_52090856 | 0.10 |
ENSDART00000155308
|
FO704712.1
|
ENSDARG00000063300 |
| chr11_+_31295037 | 0.09 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr11_+_36215690 | 0.08 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr13_-_35716712 | 0.07 |
ENSDART00000170106
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr21_+_30256863 | 0.07 |
ENSDART00000007412
|
zmat2
|
zinc finger, matrin-type 2 |
| chr15_-_46799699 | 0.06 |
|
|
|
| chr19_+_43124105 | 0.05 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr20_+_29685331 | 0.02 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr3_+_23573053 | 0.01 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr19_-_25842461 | 0.01 |
|
|
|
| chr16_+_10666588 | 0.01 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr22_-_8479330 | 0.00 |
ENSDART00000140146
ENSDART00000136891 |
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr8_-_41271615 | 0.00 |
ENSDART00000075491
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.8 | 2.5 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.7 | 2.7 | GO:0046100 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.6 | 2.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 8.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.4 | 1.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 1.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.3 | 0.9 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 1.2 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.2 | 1.4 | GO:0060296 | regulation of cilium beat frequency(GO:0003356) regulation of translational termination(GO:0006449) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 4.1 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.2 | 1.0 | GO:0046685 | regulation of myofibril size(GO:0014881) response to arsenic-containing substance(GO:0046685) |
| 0.2 | 2.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 1.1 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.2 | 0.7 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 0.5 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 0.5 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 1.8 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.2 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.7 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.7 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 2.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.3 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 3.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 1.1 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 4.5 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.7 | GO:0060415 | muscle tissue morphogenesis(GO:0060415) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.7 | 3.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.6 | 2.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 1.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 2.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.3 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.5 | 2.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.5 | 1.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.5 | 4.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.4 | 8.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 2.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 1.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 1.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 2.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 1.3 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.2 | 5.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 0.7 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 2.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.8 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 3.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.0 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 2.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 2.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 4.5 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 2.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 2.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 2.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |