Project
DANIO-CODE
Navigation
Downloads
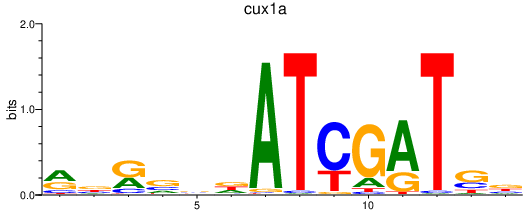
Results for cux1a
Z-value: 0.76
Transcription factors associated with cux1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux1a
|
ENSDARG00000078459 | cut-like homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux1a | dr10_dc_chr10_-_33212897_33213007 | -0.67 | 4.6e-03 | Click! |
Activity profile of cux1a motif
Sorted Z-values of cux1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of cux1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_38898811 | 1.75 |
ENSDART00000131347
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr16_-_39291647 | 1.62 |
ENSDART00000171342
|
tmem42a
|
transmembrane protein 42a |
| chr15_-_9443212 | 1.59 |
ENSDART00000177158
|
sacs
|
sacsin molecular chaperone |
| chr24_+_41991604 | 1.57 |
|
|
|
| chr17_-_7635061 | 1.54 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr14_-_6738260 | 1.39 |
ENSDART00000171792
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr6_+_38898605 | 1.39 |
ENSDART00000029930
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr2_+_58943872 | 1.29 |
ENSDART00000158860
ENSDART00000067736 |
stk11
|
serine/threonine kinase 11 |
| chr21_-_26655258 | 1.26 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr8_-_29842489 | 1.19 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr19_-_28247275 | 1.18 |
|
|
|
| chr3_-_36940649 | 1.05 |
ENSDART00000110716
ENSDART00000160233 |
tubg1
|
tubulin, gamma 1 |
| chr25_-_24440001 | 1.03 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr22_+_21491685 | 0.98 |
ENSDART00000136374
|
mier2
|
mesoderm induction early response 1, family member 2 |
| chr2_-_38223813 | 0.98 |
ENSDART00000137395
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr23_-_3778250 | 0.93 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr2_-_32530224 | 0.92 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr17_+_22291546 | 0.88 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr23_+_3778066 | 0.87 |
ENSDART00000141880
|
smim29
|
small integral membrane protein 29 |
| chr3_-_30754236 | 0.87 |
ENSDART00000109104
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr13_-_4535689 | 0.82 |
ENSDART00000023803
|
c1d
|
C1D nuclear receptor corepressor |
| chr16_-_25465777 | 0.77 |
ENSDART00000086375
|
adnp2a
|
ADNP homeobox 2a |
| chr21_+_33216466 | 0.74 |
ENSDART00000163808
|
ndst1b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1b |
| chr12_+_10078043 | 0.74 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr9_+_42656327 | 0.73 |
|
|
|
| chr13_-_22831005 | 0.71 |
ENSDART00000143112
|
tspan15
|
tetraspanin 15 |
| chr21_-_19883010 | 0.71 |
ENSDART00000137307
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_-_30022268 | 0.69 |
ENSDART00000159065
|
nucb1
|
nucleobindin 1 |
| chr4_-_30690750 | 0.64 |
ENSDART00000160757
|
CU302413.2
|
ENSDARG00000107562 |
| chr13_-_22831037 | 0.63 |
ENSDART00000057641
|
tspan15
|
tetraspanin 15 |
| chr8_+_16722505 | 0.62 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr11_+_16017857 | 0.58 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_-_56465369 | 0.56 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr7_-_37908695 | 0.55 |
|
|
|
| chr5_-_26293054 | 0.55 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr2_-_38223766 | 0.53 |
ENSDART00000137395
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr8_-_29842274 | 0.51 |
ENSDART00000086044
ENSDART00000167487 |
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr20_-_38030250 | 0.48 |
ENSDART00000142567
|
angel2
|
angel homolog 2 (Drosophila) |
| chr20_-_54285455 | 0.48 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr20_-_18836698 | 0.43 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr3_-_30754325 | 0.41 |
ENSDART00000109104
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr23_+_3778116 | 0.39 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr3_-_30022354 | 0.37 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr18_+_45574464 | 0.33 |
|
|
|
| chr3_-_11861502 | 0.32 |
ENSDART00000018159
|
ENSDARG00000011609
|
ENSDARG00000011609 |
| chr11_-_16017396 | 0.30 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr22_-_25051004 | 0.29 |
ENSDART00000131811
|
csrp2bp
|
csrp2 binding protein |
| chr15_+_44514665 | 0.28 |
|
|
|
| chr18_+_5637539 | 0.25 |
|
|
|
| chr8_-_29842822 | 0.25 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr16_-_13990964 | 0.23 |
ENSDART00000090221
ENSDART00000135099 |
cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| KN150232v1_+_13079 | 0.20 |
ENSDART00000174701
|
CABZ01084889.1
|
ENSDARG00000107269 |
| chr22_-_25051056 | 0.18 |
ENSDART00000131811
|
csrp2bp
|
csrp2 binding protein |
| chr2_+_58943688 | 0.18 |
ENSDART00000158860
ENSDART00000067736 |
stk11
|
serine/threonine kinase 11 |
| chr3_-_11861438 | 0.17 |
ENSDART00000018159
|
ENSDARG00000011609
|
ENSDARG00000011609 |
| chr25_-_5955137 | 0.15 |
ENSDART00000104755
|
cpeb1a
|
cytoplasmic polyadenylation element binding protein 1a |
| chr18_+_44709832 | 0.14 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr15_-_7622212 | 0.11 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr23_-_3778284 | 0.09 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr5_+_41396720 | 0.08 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr21_-_13565401 | 0.08 |
ENSDART00000151547
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr16_+_39292206 | 0.08 |
ENSDART00000127319
ENSDART00000102510 |
zgc:77056
|
zgc:77056 |
| chr7_+_29592486 | 0.07 |
ENSDART00000134068
|
tln2a
|
talin 2a |
| chr7_+_29592666 | 0.06 |
ENSDART00000134068
|
tln2a
|
talin 2a |
| chr16_-_24646282 | 0.06 |
ENSDART00000126274
|
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr20_+_32503118 | 0.04 |
ENSDART00000018640
|
snx3
|
sorting nexin 3 |
| chr3_-_30948297 | 0.04 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr8_-_29842345 | 0.04 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr24_-_37596370 | 0.03 |
ENSDART00000162538
|
cluap1
|
clusterin associated protein 1 |
| chr10_-_11439269 | 0.02 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr24_-_3375610 | 0.01 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr7_-_56465497 | 0.01 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr7_+_29592358 | 0.00 |
ENSDART00000167286
|
tln2a
|
talin 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.3 | 3.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 1.0 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 2.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.7 | GO:0030210 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) sulfation(GO:0051923) |
| 0.0 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.4 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.3 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.5 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.8 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 3.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 2.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.0 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.5 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |