Project
DANIO-CODE
Navigation
Downloads
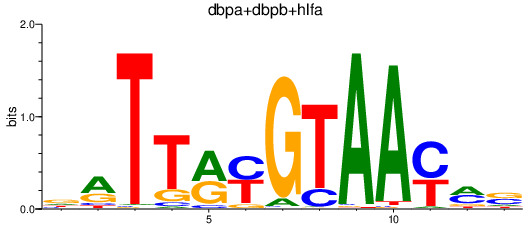
Results for dbpa+dbpb+hlfa
Z-value: 0.97
Transcription factors associated with dbpa+dbpb+hlfa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dbpb
|
ENSDARG00000057652 | D site albumin promoter binding protein b |
|
dbpa
|
ENSDARG00000063014 | D site albumin promoter binding protein a |
|
hlfa
|
ENSDARG00000074752 | HLF transcription factor, PAR bZIP family member a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dbpb | dr10_dc_chr16_-_13723295_13723320 | -0.89 | 3.0e-06 | Click! |
| dbpa | dr10_dc_chr19_-_10277398_10277421 | 0.39 | 1.3e-01 | Click! |
| hlfa | dr10_dc_chr3_-_11657881_11657973 | -0.03 | 9.2e-01 | Click! |
Activity profile of dbpa+dbpb+hlfa motif
Sorted Z-values of dbpa+dbpb+hlfa motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dbpa+dbpb+hlfa
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_7937128 | 3.64 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr1_-_35196892 | 2.94 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr16_-_45103102 | 2.03 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_-_44724371 | 2.01 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr22_+_15946773 | 1.98 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr24_-_39884167 | 1.76 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr6_+_16341787 | 1.75 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr3_-_21217799 | 1.68 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr8_+_1122000 | 1.65 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr16_+_17705704 | 1.64 |
|
|
|
| chr10_+_39140943 | 1.62 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr21_-_4842528 | 1.62 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr16_+_23482744 | 1.59 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr3_-_31672763 | 1.56 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr4_-_12726432 | 1.49 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr19_-_22914516 | 1.45 |
|
|
|
| chr24_+_38783264 | 1.42 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr25_+_7660590 | 1.42 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr12_-_5085227 | 1.39 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr8_+_7937320 | 1.33 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr17_-_52020689 | 1.32 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr18_+_618393 | 1.31 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr18_-_16933517 | 1.30 |
|
|
|
| chr16_+_46145286 | 1.28 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr7_-_38521262 | 1.28 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr13_-_35908743 | 1.27 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr21_+_25588388 | 1.21 |
ENSDART00000134678
|
ENSDARG00000078256
|
ENSDARG00000078256 |
| chr4_-_22617898 | 1.19 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr20_-_35343242 | 1.19 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr9_+_33979089 | 1.18 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr5_-_36349284 | 1.18 |
ENSDART00000047269
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr3_+_29583029 | 1.17 |
|
|
|
| chr13_-_18064929 | 1.17 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr4_-_12915292 | 1.17 |
ENSDART00000067135
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr23_+_22977031 | 1.16 |
|
|
|
| chr6_+_8895437 | 1.16 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr10_-_36864268 | 1.15 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr19_-_23037220 | 1.14 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr4_-_5294280 | 1.13 |
ENSDART00000178921
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr21_-_39013447 | 1.12 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr19_+_5073991 | 1.11 |
ENSDART00000151050
|
ENSDARG00000011076
|
ENSDARG00000011076 |
| chr16_+_34569479 | 1.06 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr5_+_18837448 | 1.01 |
|
|
|
| chr8_+_50498398 | 0.97 |
|
|
|
| chr18_+_24575639 | 0.96 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr4_+_22757046 | 0.94 |
|
|
|
| chr2_+_289138 | 0.93 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr19_-_44459390 | 0.93 |
ENSDART00000125168
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr15_+_32853646 | 0.93 |
ENSDART00000167515
|
postnb
|
periostin, osteoblast specific factor b |
| chr1_-_9387290 | 0.92 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr17_-_31327210 | 0.91 |
ENSDART00000154013
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr14_+_30568242 | 0.87 |
|
|
|
| chr5_-_43334777 | 0.85 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr24_-_32750010 | 0.85 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr8_+_8673307 | 0.84 |
ENSDART00000110854
|
elk1
|
ELK1, member of ETS oncogene family |
| chr5_+_69036145 | 0.84 |
ENSDART00000174403
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr17_-_17744885 | 0.84 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr6_-_14162846 | 0.84 |
ENSDART00000100762
|
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr19_+_16111610 | 0.80 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr2_+_1825714 | 0.80 |
ENSDART00000148624
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr20_-_35343057 | 0.79 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr23_+_4083697 | 0.79 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr11_+_23695123 | 0.79 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr17_-_8111181 | 0.77 |
ENSDART00000149873
ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr23_-_27418761 | 0.75 |
ENSDART00000022042
|
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr6_+_16342020 | 0.74 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr19_+_341775 | 0.74 |
ENSDART00000151013
|
ensaa
|
endosulfine alpha a |
| chr5_-_68439806 | 0.72 |
ENSDART00000168213
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr6_+_19965515 | 0.72 |
ENSDART00000151240
|
si:dkey-156n14.3
|
si:dkey-156n14.3 |
| chr17_-_16079935 | 0.72 |
|
|
|
| chr19_+_9113356 | 0.68 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr18_+_26916897 | 0.67 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr11_+_25266623 | 0.67 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr8_+_41274117 | 0.67 |
ENSDART00000084439
|
olfml2a
|
olfactomedin-like 2A |
| chr16_-_20488351 | 0.65 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr15_+_6070537 | 0.65 |
ENSDART00000123797
|
pcp4b
|
Purkinje cell protein 4b |
| chr10_+_10252074 | 0.64 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr7_+_69290855 | 0.64 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr25_-_211879 | 0.64 |
ENSDART00000021614
|
srpk2
|
SRSF protein kinase 2 |
| chr15_-_29415309 | 0.62 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr16_+_5508950 | 0.61 |
ENSDART00000138674
|
plecb
|
plectin b |
| chr23_+_4083748 | 0.60 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr13_-_35908796 | 0.60 |
ENSDART00000084929
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr9_+_1177938 | 0.59 |
ENSDART00000176589
|
CABZ01066921.1
|
ENSDARG00000106418 |
| chr14_+_1152700 | 0.59 |
ENSDART00000125203
|
hopx
|
HOP homeobox |
| KN150708v1_-_31573 | 0.58 |
ENSDART00000160040
|
slc16a6a
|
solute carrier family 16, member 6a |
| chr3_+_25247071 | 0.57 |
|
|
|
| chr15_-_21003820 | 0.56 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr17_-_14956738 | 0.55 |
|
|
|
| chr24_-_38782898 | 0.55 |
ENSDART00000155542
|
FP102170.1
|
ENSDARG00000097123 |
| chr10_+_32949485 | 0.55 |
|
|
|
| chr9_+_32490721 | 0.55 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr23_-_33434838 | 0.53 |
ENSDART00000031638
|
slc48a1a
|
solute carrier family 48 (heme transporter), member 1a |
| chr4_-_5293599 | 0.53 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr16_-_31670211 | 0.53 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr3_-_28534446 | 0.52 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr25_-_1989785 | 0.51 |
ENSDART00000156925
|
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr10_+_18953671 | 0.50 |
ENSDART00000135786
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr21_-_91068 | 0.48 |
|
|
|
| chr14_-_32543646 | 0.48 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr2_+_50126491 | 0.46 |
ENSDART00000144060
|
rpl37
|
ribosomal protein L37 |
| chr2_-_54662966 | 0.45 |
ENSDART00000162403
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr9_-_35260503 | 0.45 |
ENSDART00000125536
|
appb
|
amyloid beta (A4) precursor protein b |
| chr16_-_44575942 | 0.44 |
|
|
|
| chr4_+_25296408 | 0.42 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr6_+_7936792 | 0.42 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr4_+_22617563 | 0.41 |
|
|
|
| chr7_-_23577984 | 0.41 |
|
|
|
| chr8_-_29842345 | 0.41 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_+_50432868 | 0.39 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr17_+_24018420 | 0.39 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr23_+_34037058 | 0.38 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr11_+_36984616 | 0.38 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr4_+_25296376 | 0.36 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr10_-_24401876 | 0.35 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr22_+_10630419 | 0.35 |
ENSDART00000105835
|
tusc2b
|
tumor suppressor candidate 2b |
| chr3_+_24564249 | 0.35 |
ENSDART00000111769
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr6_+_7376428 | 0.33 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr9_-_21427331 | 0.33 |
ENSDART00000102143
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr11_+_16018035 | 0.31 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr15_+_28370102 | 0.30 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr24_+_28481885 | 0.30 |
ENSDART00000079770
ENSDART00000147063 |
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr5_-_19691538 | 0.30 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr9_+_1177862 | 0.30 |
ENSDART00000176589
|
CABZ01066921.1
|
ENSDARG00000106418 |
| chr21_+_7627327 | 0.30 |
ENSDART00000172174
|
agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr12_-_20251183 | 0.30 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr22_+_20695983 | 0.29 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr20_-_45757288 | 0.29 |
ENSDART00000124582
ENSDART00000100290 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr10_-_25629613 | 0.29 |
ENSDART00000131640
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr17_+_25815491 | 0.29 |
ENSDART00000164160
|
acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr7_+_10346939 | 0.28 |
|
|
|
| chr5_+_25521198 | 0.28 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr22_-_18466781 | 0.27 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr2_+_2295529 | 0.26 |
ENSDART00000127286
|
si:ch73-140j24.4
|
si:ch73-140j24.4 |
| chr17_-_31290969 | 0.26 |
|
|
|
| chr20_-_20455731 | 0.26 |
ENSDART00000170488
ENSDART00000047580 |
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr2_+_49846085 | 0.26 |
|
|
|
| chr21_+_4345150 | 0.25 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr16_-_33151504 | 0.24 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr15_-_2790691 | 0.23 |
ENSDART00000102096
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr25_-_10992022 | 0.22 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr16_+_17706003 | 0.18 |
|
|
|
| chr6_+_50452229 | 0.17 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr17_+_34168095 | 0.17 |
|
|
|
| chr12_-_26291790 | 0.16 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr22_+_15947127 | 0.16 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr9_+_24344001 | 0.15 |
|
|
|
| chr15_+_31956131 | 0.15 |
|
|
|
| chr25_+_15257730 | 0.14 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr20_-_54942216 | 0.13 |
|
|
|
| chr16_-_20488542 | 0.12 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr13_-_25643419 | 0.12 |
ENSDART00000145640
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr17_-_20959288 | 0.11 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr12_-_31369772 | 0.07 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr5_+_31562242 | 0.07 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr22_-_17571993 | 0.03 |
|
|
|
| chr16_+_6789780 | 0.02 |
ENSDART00000149742
|
znf407
|
zinc finger protein 407 |
| chr6_+_50452131 | 0.02 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr2_+_24152953 | 0.02 |
ENSDART00000131030
|
gorasp1a
|
golgi reassembly stacking protein 1a |
| chr12_-_3630622 | 0.02 |
ENSDART00000164707
|
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr13_-_25066650 | 0.01 |
|
|
|
| chr12_+_4651037 | 0.01 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr13_-_16127236 | 0.01 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr1_-_44513922 | 0.01 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr19_-_7372767 | 0.00 |
ENSDART00000137654
|
sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr6_-_32425011 | 0.00 |
ENSDART00000078908
|
usp1
|
ubiquitin specific peptidase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0042364 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) regulation of calcium-mediated signaling(GO:0050848) |
| 0.3 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 0.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 0.7 | GO:0086010 | mechanosensory behavior(GO:0007638) membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.8 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 0.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.2 | 1.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 2.0 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 0.8 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 1.4 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.2 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 0.8 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.5 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 1.3 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 1.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.1 | GO:0042129 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.1 | 4.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.1 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.9 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 2.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.7 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 1.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.2 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 2.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 1.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 2.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.3 | 1.4 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 2.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 1.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 1.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 2.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 0.8 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.8 | GO:1902387 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 1.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.8 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 2.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |