Project
DANIO-CODE
Navigation
Downloads
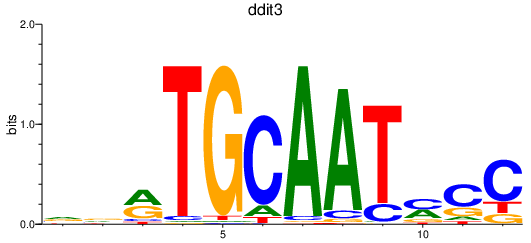
Results for ddit3
Z-value: 1.03
Transcription factors associated with ddit3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ddit3
|
ENSDARG00000059836 | DNA-damage-inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ddit3 | dr10_dc_chr6_+_58921655_58921664 | 0.85 | 2.8e-05 | Click! |
Activity profile of ddit3 motif
Sorted Z-values of ddit3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ddit3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_53286155 | 2.97 |
ENSDART00000139625
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr5_+_37378340 | 2.78 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr3_-_54959360 | 2.57 |
ENSDART00000154031
|
kank2
|
KN motif and ankyrin repeat domains 2 |
| chr17_-_32473712 | 2.50 |
ENSDART00000148455
ENSDART00000149885 |
grhl1
|
grainyhead-like transcription factor 1 |
| chr2_-_26872047 | 2.13 |
ENSDART00000138693
|
si:ch211-106k21.5
|
si:ch211-106k21.5 |
| chr14_-_5239605 | 2.01 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr23_-_42775849 | 1.97 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr7_-_11812634 | 1.96 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr3_+_36282205 | 1.95 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr13_-_33696425 | 1.93 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr7_+_55332040 | 1.90 |
ENSDART00000171736
|
trappc2l
|
trafficking protein particle complex 2-like |
| chr3_+_12613413 | 1.89 |
ENSDART00000166071
|
cyp2k16
|
cytochrome P450, family 2, subfamily K, polypeptide16 |
| chr17_+_16067769 | 1.87 |
ENSDART00000155292
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_+_9285446 | 1.78 |
ENSDART00000033663
|
rps21
|
ribosomal protein S21 |
| chr2_+_36634309 | 1.60 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr13_+_30673907 | 1.58 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr9_-_42682501 | 1.55 |
ENSDART00000048320
|
tfpia
|
tissue factor pathway inhibitor a |
| chr5_+_6393799 | 1.53 |
ENSDART00000099417
|
CABZ01041962.1
|
ENSDARG00000076082 |
| chr4_-_12862708 | 1.49 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr5_-_23301474 | 1.48 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr22_+_16511562 | 1.44 |
|
|
|
| chr25_-_31452381 | 1.41 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr4_+_15870605 | 1.39 |
ENSDART00000178674
|
BX004864.1
|
ENSDARG00000107641 |
| chr1_+_33563354 | 1.36 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr22_-_21289850 | 1.28 |
ENSDART00000135388
|
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr4_+_29209923 | 1.26 |
ENSDART00000168709
|
ENSDARG00000104041
|
ENSDARG00000104041 |
| chr5_-_56981925 | 1.25 |
|
|
|
| chr6_+_13833991 | 1.25 |
|
|
|
| chr17_+_15425559 | 1.23 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr5_-_50972677 | 1.22 |
ENSDART00000134606
|
otpb
|
orthopedia homeobox b |
| chr16_-_46697715 | 1.19 |
ENSDART00000159209
|
tmem176l.4
|
transmembrane protein 176l.4 |
| chr6_+_6640324 | 1.18 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr7_+_59445089 | 1.16 |
ENSDART00000175691
ENSDART00000179121 |
CR384075.1
|
ENSDARG00000108680 |
| chr2_-_6139895 | 1.13 |
ENSDART00000147509
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr23_-_10241885 | 1.11 |
ENSDART00000081223
ENSDART00000144280 |
krt5
|
keratin 5 |
| chr20_-_28981879 | 1.09 |
ENSDART00000134564
ENSDART00000132127 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr15_-_17933972 | 1.08 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr17_-_31678247 | 1.07 |
ENSDART00000143090
|
lin52
|
lin-52 DREAM MuvB core complex component |
| chr24_-_4117031 | 1.05 |
ENSDART00000147849
|
klf6a
|
Kruppel-like factor 6a |
| chr2_+_53253623 | 1.01 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr13_+_44720106 | 1.01 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr10_+_43360137 | 0.98 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr10_+_29378834 | 0.98 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr16_-_543957 | 0.97 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr23_-_3815492 | 0.94 |
ENSDART00000131536
|
hmga1a
|
high mobility group AT-hook 1a |
| chr16_-_17350056 | 0.94 |
ENSDART00000079497
|
emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr7_+_58007815 | 0.93 |
ENSDART00000179306
|
CR354540.2
|
ENSDARG00000106881 |
| chr11_-_11925832 | 0.88 |
|
|
|
| chr5_-_20527105 | 0.88 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr15_+_36329506 | 0.86 |
ENSDART00000099501
|
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr9_+_28301359 | 0.86 |
|
|
|
| chr6_+_43236843 | 0.82 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr8_-_23762309 | 0.80 |
ENSDART00000141879
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr6_-_8001756 | 0.79 |
ENSDART00000151483
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr4_+_5363065 | 0.79 |
|
|
|
| chr22_-_28922834 | 0.78 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr4_+_28860191 | 0.78 |
ENSDART00000110358
ENSDART00000162069 |
znf1059
|
zinc finger protein 1059 |
| chr8_-_11512545 | 0.78 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr22_-_16971799 | 0.76 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr5_-_68439806 | 0.75 |
ENSDART00000168213
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr20_-_32429358 | 0.75 |
ENSDART00000062982
|
foxo3b
|
forkhead box O3b |
| chr23_-_12488794 | 0.74 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr23_+_18176720 | 0.74 |
ENSDART00000010270
|
mfsd4ab
|
major facilitator superfamily domain containing 4Ab |
| chr16_+_21894717 | 0.73 |
ENSDART00000172504
|
si:ch211-154o6.3
|
si:ch211-154o6.3 |
| chr19_-_22182031 | 0.72 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr10_-_43200499 | 0.72 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr13_+_13562712 | 0.70 |
ENSDART00000110509
ENSDART00000169265 |
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr4_-_44105748 | 0.69 |
ENSDART00000150621
|
znf1137
|
zinc finger protein 1137 |
| chr19_+_9085377 | 0.68 |
ENSDART00000091443
|
ENSDARG00000062900
|
ENSDARG00000062900 |
| chr19_-_9793494 | 0.67 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr5_-_50972592 | 0.67 |
ENSDART00000134606
|
otpb
|
orthopedia homeobox b |
| chr7_-_18629056 | 0.66 |
ENSDART00000021502
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr25_+_34557335 | 0.66 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr9_-_22324699 | 0.64 |
ENSDART00000175417
ENSDART00000101902 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr23_-_14317229 | 0.64 |
|
|
|
| chr23_+_34060851 | 0.64 |
ENSDART00000037501
|
fam50a
|
family with sequence similarity 50, member A |
| chr16_+_32795616 | 0.62 |
ENSDART00000136759
|
prdm13
|
PR domain containing 13 |
| chr22_+_5657904 | 0.60 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr5_+_37184963 | 0.60 |
ENSDART00000053511
|
myo1ca
|
myosin Ic, paralog a |
| chr22_-_10612143 | 0.59 |
ENSDART00000064772
|
cyb561d2
|
cytochrome b561 family, member D2 |
| chr22_+_685232 | 0.59 |
|
|
|
| chr1_+_13661799 | 0.58 |
ENSDART00000064468
|
crmp1
|
collapsin response mediator protein 1 |
| chr2_+_13373937 | 0.58 |
ENSDART00000142649
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr2_+_10148323 | 0.56 |
ENSDART00000158214
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr3_-_26073676 | 0.55 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr21_+_22792068 | 0.54 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr23_+_14058972 | 0.54 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr7_-_22519196 | 0.53 |
ENSDART00000129919
|
ENSDARG00000091111
|
ENSDARG00000091111 |
| chr20_+_3095763 | 0.52 |
ENSDART00000133435
|
CEP170B
|
centrosomal protein 170B |
| chr23_-_20503270 | 0.51 |
ENSDART00000104334
|
taf13
|
TATA-box binding protein associated factor 13 |
| chr1_+_18272833 | 0.47 |
ENSDART00000132379
|
limch1a
|
LIM and calponin homology domains 1a |
| chr21_+_11792666 | 0.47 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr1_+_44301612 | 0.47 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr23_-_27681694 | 0.46 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr14_+_26498814 | 0.46 |
ENSDART00000137695
|
klhl4
|
kelch-like family member 4 |
| chr15_-_16387549 | 0.45 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr5_-_70688185 | 0.45 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr2_+_36634251 | 0.44 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr16_+_38709412 | 0.44 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr13_-_21529695 | 0.44 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr1_+_19376127 | 0.42 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr10_+_15819085 | 0.42 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr4_+_14928134 | 0.41 |
ENSDART00000019647
|
psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr25_+_33364224 | 0.41 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| KN150442v1_+_26452 | 0.41 |
|
|
|
| chr23_-_28766019 | 0.40 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr1_+_44044807 | 0.39 |
ENSDART00000133926
ENSDART00000049701 |
p2rx3b
|
purinergic receptor P2X, ligand-gated ion channel, 3b |
| chr4_-_5362973 | 0.37 |
ENSDART00000150697
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr8_-_45394216 | 0.37 |
ENSDART00000113837
|
mak16
|
MAK16 homolog (S. cerevisiae) |
| chr15_-_29420984 | 0.36 |
ENSDART00000127795
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr2_+_17116697 | 0.36 |
|
|
|
| chr18_+_36825511 | 0.35 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr10_+_44853121 | 0.35 |
ENSDART00000167952
|
scarb1
|
scavenger receptor class B, member 1 |
| chr14_-_23503974 | 0.35 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr2_+_10148394 | 0.34 |
ENSDART00000158214
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr8_+_17803229 | 0.34 |
ENSDART00000175436
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr5_-_40508107 | 0.34 |
|
|
|
| chr19_-_32178232 | 0.33 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr17_+_794430 | 0.32 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr21_-_2247380 | 0.32 |
ENSDART00000171417
|
si:ch73-299h12.3
|
si:ch73-299h12.3 |
| chr23_+_45876447 | 0.31 |
ENSDART00000169521
ENSDART00000162915 |
dclk2b
|
doublecortin-like kinase 2b |
| chr9_+_28301214 | 0.31 |
|
|
|
| chr3_-_26073923 | 0.31 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr25_-_34557204 | 0.30 |
|
|
|
| chr3_-_28297082 | 0.29 |
ENSDART00000151546
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr25_+_19901741 | 0.28 |
ENSDART00000026401
|
ENSDARG00000004577
|
ENSDARG00000004577 |
| chr8_+_31110356 | 0.28 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr8_+_23172960 | 0.28 |
ENSDART00000028946
|
tpd52l2a
|
tumor protein D52-like 2a |
| chr1_+_2565161 | 0.28 |
ENSDART00000152350
|
gpc6a
|
glypican 6a |
| chr21_+_11375961 | 0.27 |
ENSDART00000158936
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr23_+_24010310 | 0.27 |
ENSDART00000171029
|
ptpn11b
|
protein tyrosine phosphatase, non-receptor type 11, b |
| chr2_-_30710221 | 0.27 |
ENSDART00000090292
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr18_-_13088462 | 0.26 |
ENSDART00000088908
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr7_-_55331967 | 0.25 |
ENSDART00000149478
|
galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr21_+_4916179 | 0.25 |
ENSDART00000024199
|
atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr2_-_16669661 | 0.25 |
|
|
|
| chr14_+_7585570 | 0.24 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr2_-_45301575 | 0.22 |
ENSDART00000109890
|
capn10
|
calpain 10 |
| chr6_-_7937201 | 0.22 |
|
|
|
| chr20_+_23184685 | 0.22 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr2_-_14833381 | 0.21 |
|
|
|
| chr5_+_27671064 | 0.20 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr6_+_42340724 | 0.20 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr15_+_24811157 | 0.20 |
ENSDART00000032306
|
ywhae1
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 |
| chr20_-_4635674 | 0.20 |
|
|
|
| chr3_+_19849761 | 0.19 |
|
|
|
| chr10_+_26554189 | 0.19 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr4_+_2551928 | 0.19 |
ENSDART00000103371
ENSDART00000171405 |
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr7_-_18629119 | 0.19 |
ENSDART00000021502
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr10_+_26554237 | 0.17 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr4_+_56997555 | 0.17 |
|
|
|
| chr18_-_13088412 | 0.17 |
|
|
|
| chr21_-_97746 | 0.16 |
|
|
|
| chr6_-_8001792 | 0.13 |
ENSDART00000151483
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr21_-_21336603 | 0.13 |
ENSDART00000079629
|
ppm1nb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Nb (putative) |
| chr8_-_8307383 | 0.13 |
|
|
|
| chr2_-_14833347 | 0.13 |
|
|
|
| chr6_-_10505151 | 0.13 |
ENSDART00000150942
|
sp3b
|
Sp3b transcription factor |
| chr22_-_37091982 | 0.13 |
|
|
|
| chr9_-_25370657 | 0.13 |
ENSDART00000166684
|
esd
|
esterase D/formylglutathione hydrolase |
| chr3_-_19968543 | 0.12 |
ENSDART00000055745
ENSDART00000139902 |
sepw2a
|
selenoprotein W, 2a |
| chr17_-_45140696 | 0.11 |
|
|
|
| chr24_+_35173816 | 0.09 |
|
|
|
| chr8_-_10978942 | 0.08 |
ENSDART00000064066
|
bcas2
|
BCAS2, pre-mRNA processing factor |
| chr13_-_34732375 | 0.08 |
|
|
|
| chr7_+_16978182 | 0.08 |
|
|
|
| chr13_+_18235298 | 0.07 |
ENSDART00000133057
ENSDART00000111129 |
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr14_+_7585937 | 0.07 |
ENSDART00000063837
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr20_+_31371853 | 0.06 |
|
|
|
| chr7_-_65913414 | 0.06 |
ENSDART00000145843
|
pth1b
|
parathyroid hormone 1b |
| chr7_+_39353697 | 0.02 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr19_-_24969483 | 0.01 |
|
|
|
| chr2_-_51362397 | 0.01 |
ENSDART00000167172
|
THEM6
|
thioesterase superfamily member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0043691 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) reverse cholesterol transport(GO:0043691) positive regulation of steroid metabolic process(GO:0045940) |
| 0.9 | 2.6 | GO:0071305 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) cellular response to vitamin D(GO:0071305) |
| 0.8 | 2.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.6 | 2.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 3.0 | GO:0072132 | mesenchyme morphogenesis(GO:0072132) |
| 0.3 | 1.8 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 0.7 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.2 | 2.0 | GO:0008406 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 0.7 | GO:0002832 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.2 | 0.5 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 1.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.6 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 1.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:0003433 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.9 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.4 | GO:1903307 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.1 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:0071387 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 1.0 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 1.6 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.3 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.0 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.2 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 2.0 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.6 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 3.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.2 | GO:0098594 | mucin granule(GO:0098594) |
| 0.1 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 8.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.1 | GO:0071814 | lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.4 | 2.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.6 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.2 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 2.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.7 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0004931 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 2.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 3.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |