Project
DANIO-CODE
Navigation
Downloads
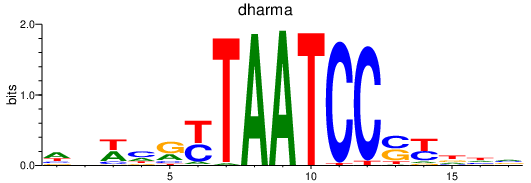
Results for dharma
Z-value: 0.40
Transcription factors associated with dharma
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dharma
|
ENSDARG00000040955 | dharma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dharma | dr10_dc_chr15_-_29079613_29079638 | -0.40 | 1.3e-01 | Click! |
Activity profile of dharma motif
Sorted Z-values of dharma motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dharma
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_28099284 | 0.42 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr23_-_28099359 | 0.41 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr12_+_11042472 | 0.41 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr23_-_28099391 | 0.38 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr23_-_28099235 | 0.33 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr10_+_32160464 | 0.32 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr14_+_34626233 | 0.31 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr23_+_20779487 | 0.31 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr3_+_22962233 | 0.30 |
ENSDART00000142884
|
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr9_+_31265296 | 0.30 |
ENSDART00000132478
|
BX470183.1
|
ENSDARG00000095476 |
| chr16_+_19731017 | 0.27 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr21_-_19293600 | 0.26 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr3_-_14348442 | 0.25 |
ENSDART00000172102
|
elof1
|
ELF1 homolog, elongation factor 1 |
| chr15_-_5575675 | 0.24 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr14_-_25688444 | 0.24 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr16_+_19730958 | 0.23 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr12_+_11042439 | 0.23 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr9_+_26218474 | 0.23 |
ENSDART00000177394
|
CR391949.1
|
ENSDARG00000107108 |
| chr4_+_11054808 | 0.22 |
ENSDART00000140362
|
ccdc59
|
coiled-coil domain containing 59 |
| chr18_-_15582896 | 0.22 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr18_+_17765358 | 0.21 |
ENSDART00000137239
|
si:ch211-216l23.2
|
si:ch211-216l23.2 |
| chr18_-_5235925 | 0.20 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr21_-_2322963 | 0.19 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr14_-_952510 | 0.19 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr17_+_25407644 | 0.19 |
ENSDART00000149557
|
srrm1
|
serine/arginine repetitive matrix 1 |
| chr23_+_44828346 | 0.18 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr12_-_954375 | 0.18 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr5_+_8642839 | 0.18 |
ENSDART00000003273
|
rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr15_+_15580544 | 0.17 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr16_+_19730756 | 0.17 |
ENSDART00000112894
|
sp8b
|
sp8 transcription factor b |
| chr16_+_42925950 | 0.17 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr15_-_8215831 | 0.17 |
ENSDART00000155381
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr21_+_44749057 | 0.17 |
ENSDART00000084780
|
rnf121
|
ring finger protein 121 |
| chr2_+_3671135 | 0.17 |
ENSDART00000113395
|
SERP1
|
stress associated endoplasmic reticulum protein 1 |
| chr12_-_13866811 | 0.16 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr1_-_44227620 | 0.16 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr10_-_11439269 | 0.16 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr16_+_19730820 | 0.15 |
ENSDART00000079201
|
sp8b
|
sp8 transcription factor b |
| chr7_+_71096788 | 0.15 |
ENSDART00000161871
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr7_+_38123920 | 0.15 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr1_-_44476084 | 0.14 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr1_+_50835265 | 0.14 |
ENSDART00000162226
|
meis1a
|
Meis homeobox 1 a |
| chr5_-_23301474 | 0.14 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr25_+_35771748 | 0.14 |
|
|
|
| chr16_+_54729975 | 0.14 |
|
|
|
| chr21_+_30256863 | 0.14 |
ENSDART00000007412
|
zmat2
|
zinc finger, matrin-type 2 |
| chr3_+_1440282 | 0.14 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr19_+_1106964 | 0.14 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr8_-_43568078 | 0.13 |
ENSDART00000051139
ENSDART00000160289 |
ncor2
|
nuclear receptor corepressor 2 |
| chr14_+_6834563 | 0.13 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr17_-_6191539 | 0.13 |
ENSDART00000081707
|
ENSDARG00000058775
|
ENSDARG00000058775 |
| chr14_+_34626191 | 0.13 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr24_-_1014318 | 0.12 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr1_+_13779755 | 0.12 |
|
|
|
| chr17_-_33462243 | 0.12 |
ENSDART00000131807
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr21_-_20305406 | 0.12 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr20_+_13885641 | 0.12 |
ENSDART00000142999
|
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr3_+_38497816 | 0.12 |
|
|
|
| chr14_-_6835186 | 0.12 |
ENSDART00000172020
|
trpt1
|
tRNA phosphotransferase 1 |
| chr16_+_11260802 | 0.12 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr20_-_46638031 | 0.11 |
ENSDART00000060685
|
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr23_-_28368224 | 0.11 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr6_-_17699493 | 0.11 |
ENSDART00000154180
|
CR388029.1
|
ENSDARG00000097214 |
| KN149926v1_+_1959 | 0.10 |
|
|
|
| chr9_+_3164027 | 0.10 |
ENSDART00000160567
ENSDART00000161216 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr5_-_68943821 | 0.10 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr21_-_6540681 | 0.10 |
|
|
|
| chr11_+_18823629 | 0.10 |
|
|
|
| chr2_-_9946238 | 0.10 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr5_+_35729588 | 0.10 |
|
|
|
| chr15_-_8215797 | 0.10 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr3_-_13359125 | 0.10 |
ENSDART00000166832
|
slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr5_-_43160243 | 0.09 |
|
|
|
| chr11_+_15478585 | 0.09 |
ENSDART00000163289
ENSDART00000066033 |
gdf11
|
growth differentiation factor 11 |
| chr15_-_15532980 | 0.09 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr13_-_39821399 | 0.09 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr18_+_38210277 | 0.09 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr15_-_5575746 | 0.09 |
ENSDART00000130297
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr16_+_9635202 | 0.09 |
ENSDART00000148699
|
BX324207.1
|
ENSDARG00000095943 |
| chr14_+_50700280 | 0.08 |
ENSDART00000174722
|
CU929193.1
|
ENSDARG00000106699 |
| chr10_+_7296895 | 0.08 |
ENSDART00000161506
|
fut10
|
fucosyltransferase 10 |
| chr5_-_29852219 | 0.08 |
|
|
|
| chr14_+_8332180 | 0.08 |
ENSDART00000123506
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr3_+_45375639 | 0.08 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr11_-_40414131 | 0.08 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr11_-_3592968 | 0.08 |
ENSDART00000085855
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr3_-_29760674 | 0.07 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| chr5_+_56772028 | 0.07 |
ENSDART00000097395
|
prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr3_-_36339966 | 0.07 |
ENSDART00000176547
|
ppl
|
periplakin |
| chr11_-_21424553 | 0.07 |
ENSDART00000045391
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_-_32404106 | 0.07 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr23_-_41929027 | 0.07 |
ENSDART00000131235
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr14_-_25637853 | 0.07 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr7_+_66660781 | 0.07 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr8_-_3354350 | 0.07 |
ENSDART00000169430
ENSDART00000167187 |
FUT9 (1 of many)
|
fucosyltransferase 9 |
| chr2_-_36845638 | 0.07 |
|
|
|
| chr10_-_5016472 | 0.07 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr7_-_41601001 | 0.07 |
ENSDART00000174258
|
zgc:92818
|
zgc:92818 |
| chr24_-_41114517 | 0.06 |
ENSDART00000162783
|
CU633479.1
|
ENSDARG00000098266 |
| chr25_-_34616837 | 0.06 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr16_+_29651305 | 0.06 |
ENSDART00000124232
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr23_-_41929112 | 0.06 |
ENSDART00000131235
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr4_+_22759177 | 0.06 |
ENSDART00000146272
|
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr21_+_5528438 | 0.06 |
ENSDART00000168158
|
shroom3
|
shroom family member 3 |
| chr4_+_952596 | 0.06 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr10_-_5016621 | 0.06 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr24_+_27149156 | 0.06 |
|
|
|
| chr20_+_23769136 | 0.05 |
ENSDART00000155262
|
nek1
|
NIMA-related kinase 1 |
| chr16_-_51087813 | 0.05 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr16_-_52090856 | 0.05 |
ENSDART00000155308
|
FO704712.1
|
ENSDARG00000063300 |
| chr3_+_45375698 | 0.05 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr6_-_17699766 | 0.05 |
ENSDART00000170597
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr18_-_19016801 | 0.05 |
ENSDART00000129776
|
vwa9
|
von Willebrand factor A domain containing 9 |
| chr3_-_6968741 | 0.05 |
|
|
|
| chr5_-_51351827 | 0.05 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr2_+_9762627 | 0.04 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr25_+_34546629 | 0.04 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr14_+_7585570 | 0.04 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr4_-_11054645 | 0.04 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr8_+_28419432 | 0.04 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr21_-_21574214 | 0.04 |
ENSDART00000172333
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr7_+_17701832 | 0.04 |
ENSDART00000113120
|
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr24_-_37810565 | 0.04 |
ENSDART00000172178
|
si:ch211-231f6.6
|
si:ch211-231f6.6 |
| chr19_+_1106826 | 0.04 |
ENSDART00000165516
|
cfap20
|
cilia and flagella associated protein 20 |
| chr15_+_17912538 | 0.04 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr3_+_43125015 | 0.04 |
ENSDART00000165145
|
litaf
|
lipopolysaccharide-induced TNF factor |
| chr3_-_32190009 | 0.04 |
|
|
|
| chr24_-_3273409 | 0.03 |
|
|
|
| chr8_+_46319434 | 0.03 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr3_+_16514376 | 0.03 |
ENSDART00000013816
|
ENSDARG00000013161
|
ENSDARG00000013161 |
| chr1_-_44227583 | 0.03 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| KN149959v1_+_21015 | 0.03 |
ENSDART00000161495
|
CABZ01078608.1
|
ENSDARG00000105028 |
| chr5_-_23741221 | 0.03 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr9_+_25109771 | 0.03 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr19_+_44310050 | 0.03 |
ENSDART00000168725
ENSDART00000133628 |
ankib1a
|
ankyrin repeat and IBR domain containing 1a |
| chr22_-_21289850 | 0.03 |
ENSDART00000135388
|
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr14_-_47408279 | 0.03 |
ENSDART00000083833
|
bbs7
|
Bardet-Biedl syndrome 7 |
| chr1_-_43323814 | 0.03 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr11_+_17849528 | 0.03 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr11_-_40414239 | 0.03 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr25_-_6262259 | 0.03 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr5_-_39440105 | 0.02 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr5_+_29301998 | 0.02 |
ENSDART00000175940
|
BX284646.1
|
ENSDARG00000106174 |
| chr9_+_25109716 | 0.02 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr7_+_61693871 | 0.02 |
|
|
|
| chr1_+_39315964 | 0.02 |
ENSDART00000011727
|
ccdc149b
|
coiled-coil domain containing 149b |
| chr2_-_9946148 | 0.02 |
ENSDART00000175460
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr25_-_34616997 | 0.01 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr18_+_50528517 | 0.01 |
ENSDART00000151647
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr5_+_12894381 | 0.01 |
ENSDART00000051669
|
tctn2
|
tectonic family member 2 |
| chr5_-_68943600 | 0.01 |
ENSDART00000159851
|
mob1a
|
MOB kinase activator 1A |
| chr10_+_11439845 | 0.01 |
ENSDART00000064210
|
cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr3_+_35478909 | 0.01 |
ENSDART00000132703
|
traf7
|
TNF receptor-associated factor 7 |
| chr16_+_29651573 | 0.01 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr19_+_46717285 | 0.01 |
ENSDART00000170505
|
rprd1a
|
regulation of nuclear pre-mRNA domain containing 1A |
| chr14_-_3836967 | 0.01 |
ENSDART00000055817
|
ENSDARG00000038270
|
ENSDARG00000038270 |
| chr6_+_7263696 | 0.00 |
|
|
|
| chr6_-_39313358 | 0.00 |
|
|
|
| chr4_+_6630737 | 0.00 |
ENSDART00000158573
|
SMIM30
|
si:dkey-112e7.2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.2 | GO:2000253 | regulation of somitogenesis(GO:0014807) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.3 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 1.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.2 | GO:1903069 | neuronal action potential(GO:0019228) regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.1 | GO:0033262 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0050855 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) vitamin transmembrane transporter activity(GO:0090482) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |