Project
DANIO-CODE
Navigation
Downloads
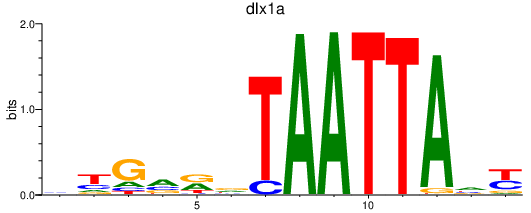
Results for dlx1a
Z-value: 0.81
Transcription factors associated with dlx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx1a
|
ENSDARG00000013125 | distal-less homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx1a | dr10_dc_chr9_+_3416651_3416654 | -0.86 | 1.8e-05 | Click! |
Activity profile of dlx1a motif
Sorted Z-values of dlx1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_24727689 | 2.39 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr11_-_6442588 | 2.18 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr18_-_40718244 | 2.12 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| KN150456v1_-_19515 | 1.97 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr10_-_21404605 | 1.92 |
ENSDART00000125167
|
avd
|
avidin |
| chr6_+_23476669 | 1.87 |
|
|
|
| chr21_-_32027717 | 1.69 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr17_+_16038358 | 1.65 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr14_-_33141111 | 1.64 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr3_+_42383724 | 1.54 |
|
|
|
| chr25_+_5845303 | 1.47 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr12_-_36046366 | 1.46 |
|
|
|
| chr19_-_20819477 | 1.42 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr7_+_35163845 | 1.40 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr21_+_25740782 | 1.40 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr12_-_33256754 | 1.40 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr4_+_4825461 | 1.38 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr5_+_36168475 | 1.38 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr19_-_20819101 | 1.35 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr24_+_19270877 | 1.33 |
|
|
|
| chr21_+_20360180 | 1.31 |
ENSDART00000003299
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr11_-_44539778 | 1.31 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr10_+_15066791 | 1.29 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr7_-_47978449 | 1.26 |
ENSDART00000127007
ENSDART00000024062 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr16_+_42567707 | 1.26 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr11_-_1524107 | 1.25 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr1_+_46474561 | 1.24 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr20_-_14218080 | 1.21 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr14_+_22159893 | 1.21 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr10_-_8088063 | 1.20 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr18_-_43890836 | 1.18 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr23_+_32174669 | 1.17 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| KN150030v1_-_22572 | 1.17 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr10_-_4641624 | 1.16 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr17_+_16038103 | 1.16 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr9_+_29737843 | 1.10 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr7_-_73613531 | 1.10 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr22_-_38321005 | 1.09 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr18_-_38787413 | 1.07 |
|
|
|
| chr13_+_33578737 | 1.07 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr11_-_2437396 | 1.05 |
|
|
|
| chr8_-_53165501 | 1.05 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_-_1571878 | 1.04 |
|
|
|
| chr19_+_5562075 | 1.04 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr11_+_17849608 | 1.03 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr19_+_770458 | 1.02 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr18_+_9755131 | 1.01 |
|
|
|
| chr22_+_17235696 | 1.01 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr21_+_6289363 | 1.00 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr6_-_40715613 | 0.99 |
ENSDART00000153702
|
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr7_-_48394268 | 0.99 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr6_+_54672873 | 0.99 |
ENSDART00000074602
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr4_+_71390178 | 0.98 |
ENSDART00000174128
|
zgc:152938
|
zgc:152938 |
| chr17_-_7575628 | 0.97 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr6_-_10676857 | 0.97 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| KN150266v1_-_68652 | 0.97 |
|
|
|
| chr12_+_22459177 | 0.96 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr1_-_51075777 | 0.94 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr13_-_28653891 | 0.93 |
ENSDART00000007741
|
ENSDARG00000013581
|
ENSDARG00000013581 |
| chr1_-_22617455 | 0.93 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr10_+_44853207 | 0.92 |
ENSDART00000169466
|
scarb1
|
scavenger receptor class B, member 1 |
| chr25_+_29219342 | 0.91 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr12_+_23691261 | 0.91 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr13_-_18564182 | 0.91 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr21_-_2273151 | 0.91 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr8_+_41003546 | 0.91 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr12_-_33256671 | 0.90 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr1_+_9443056 | 0.90 |
ENSDART00000110698
|
rbm46
|
RNA binding motif protein 46 |
| chr11_+_11319810 | 0.89 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr16_-_43061368 | 0.89 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr14_-_8634381 | 0.89 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr20_+_14218237 | 0.88 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr22_+_1837102 | 0.87 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr24_-_14446593 | 0.87 |
|
|
|
| chr11_+_44002664 | 0.87 |
|
|
|
| chr23_-_35691369 | 0.87 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr23_-_25853364 | 0.87 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr1_+_43471798 | 0.86 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr8_+_16641127 | 0.86 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr12_+_22459218 | 0.85 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_+_25302180 | 0.83 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr23_+_22857958 | 0.83 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr12_+_23691397 | 0.83 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr21_-_45323151 | 0.82 |
ENSDART00000151106
|
ENSDARG00000053404
|
ENSDARG00000053404 |
| chr13_-_10214396 | 0.82 |
ENSDART00000132231
|
AL929457.1
|
ENSDARG00000095483 |
| chr9_+_45193290 | 0.81 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr2_-_40067309 | 0.81 |
|
|
|
| chr1_+_8468971 | 0.81 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr3_-_61258158 | 0.79 |
ENSDART00000123388
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr7_-_21571924 | 0.79 |
ENSDART00000166446
|
BX005336.1
|
ENSDARG00000102693 |
| chr4_-_3340315 | 0.78 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr16_+_27837465 | 0.78 |
|
|
|
| chr14_+_15845853 | 0.78 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr13_-_37340209 | 0.78 |
|
|
|
| chr12_-_33257026 | 0.78 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr6_+_18941135 | 0.78 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr3_+_12087549 | 0.77 |
|
|
|
| chr3_-_30955799 | 0.77 |
ENSDART00000103421
|
zgc:153292
|
zgc:153292 |
| chr3_-_39912816 | 0.77 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr1_+_43471683 | 0.77 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr9_-_705001 | 0.77 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr21_-_11562388 | 0.76 |
ENSDART00000139289
|
cast
|
calpastatin |
| chr12_+_4885307 | 0.76 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr10_-_17214368 | 0.76 |
|
|
|
| chr17_-_36913213 | 0.76 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr2_-_15656155 | 0.75 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr7_+_66660882 | 0.75 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr18_+_8954512 | 0.75 |
ENSDART00000134827
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr2_+_15379961 | 0.75 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr14_-_15777250 | 0.75 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr12_+_33257120 | 0.74 |
|
|
|
| chr11_+_12754166 | 0.74 |
ENSDART00000123445
ENSDART00000163364 ENSDART00000066122 |
rtel1
|
regulator of telomere elongation helicase 1 |
| chr21_+_11686037 | 0.74 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr18_-_23847340 | 0.74 |
ENSDART00000155068
|
BX936364.1
|
ENSDARG00000097764 |
| chr14_+_17092009 | 0.73 |
ENSDART00000129838
|
rnf212
|
ring finger protein 212 |
| chr20_-_20370323 | 0.73 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr7_-_73613763 | 0.72 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr11_-_33605979 | 0.72 |
ENSDART00000171439
|
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr6_-_8156471 | 0.72 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr2_+_30800532 | 0.72 |
|
|
|
| chr22_-_29695242 | 0.72 |
|
|
|
| chr15_+_35076414 | 0.71 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr2_-_56848527 | 0.71 |
ENSDART00000164330
|
CU634008.1
|
ENSDARG00000100430 |
| chr10_-_35313462 | 0.71 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr7_+_15059782 | 0.71 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr5_-_4483970 | 0.70 |
|
|
|
| chr2_+_56534374 | 0.70 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr25_-_20932941 | 0.70 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr8_-_44247277 | 0.69 |
|
|
|
| chr12_-_33256934 | 0.69 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr6_-_12687107 | 0.68 |
ENSDART00000155360
|
CU639412.1
|
ENSDARG00000096929 |
| chr11_-_11809024 | 0.68 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr15_+_29207125 | 0.67 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr23_+_44814692 | 0.67 |
|
|
|
| chr1_+_43471754 | 0.67 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr13_+_20393886 | 0.66 |
ENSDART00000081385
|
ccdc172
|
coiled-coil domain containing 172 |
| chr23_+_44814525 | 0.66 |
|
|
|
| chr16_+_25381315 | 0.66 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr11_-_2437361 | 0.66 |
|
|
|
| KN150030v1_-_22613 | 0.66 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr25_-_8026912 | 0.65 |
|
|
|
| chr8_-_19215929 | 0.65 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr11_+_25302072 | 0.65 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr25_-_36616544 | 0.65 |
|
|
|
| chr6_+_342822 | 0.65 |
|
|
|
| chr14_-_16118942 | 0.64 |
|
|
|
| chr8_-_23591293 | 0.64 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr18_+_18011759 | 0.63 |
ENSDART00000147797
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr7_-_5253629 | 0.63 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr25_-_13394261 | 0.63 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr5_-_38570658 | 0.63 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr15_-_43402935 | 0.63 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr20_+_6545449 | 0.62 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr20_-_44377905 | 0.62 |
ENSDART00000177089
ENSDART00000176887 |
CABZ01035100.1
|
ENSDARG00000106328 |
| chr17_-_26519469 | 0.62 |
ENSDART00000155692
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr21_-_39132388 | 0.61 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr8_-_22252628 | 0.61 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr15_+_29460803 | 0.61 |
ENSDART00000155198
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr1_+_51998406 | 0.61 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr6_+_18941186 | 0.61 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr7_-_54752916 | 0.61 |
|
|
|
| chr18_+_39506453 | 0.60 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr21_-_2273243 | 0.60 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr3_+_52290252 | 0.60 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr4_+_5147577 | 0.60 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr20_+_29306863 | 0.59 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr12_+_4885394 | 0.59 |
ENSDART00000159367
|
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr10_+_35313772 | 0.59 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr13_-_30741976 | 0.59 |
ENSDART00000142535
|
CR762493.1
|
ENSDARG00000092411 |
| chr18_-_23847383 | 0.59 |
ENSDART00000155068
|
BX936364.1
|
ENSDARG00000097764 |
| chr23_+_44635770 | 0.59 |
|
|
|
| chr20_-_37910887 | 0.59 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr10_+_15066970 | 0.59 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr8_-_39788989 | 0.59 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr7_-_73016194 | 0.58 |
|
|
|
| chr19_+_770644 | 0.58 |
ENSDART00000150926
|
gstr
|
glutathione S-transferase rho |
| chr8_+_28362546 | 0.58 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr23_-_19299279 | 0.58 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_15379717 | 0.57 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr23_-_32173435 | 0.57 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr18_+_5066229 | 0.57 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr7_-_21572054 | 0.57 |
ENSDART00000166446
|
BX005336.1
|
ENSDARG00000102693 |
| chr12_-_33256599 | 0.57 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_26052785 | 0.57 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr25_-_20933145 | 0.57 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr16_+_42567668 | 0.57 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr2_-_26941084 | 0.56 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr1_-_55195566 | 0.56 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr2_-_47612319 | 0.56 |
ENSDART00000133615
|
BX085193.1
|
ENSDARG00000091979 |
| chr10_+_15066654 | 0.56 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr6_+_49724436 | 0.56 |
ENSDART00000154738
|
stx16
|
syntaxin 16 |
| chr15_+_23592828 | 0.56 |
ENSDART00000152422
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr4_-_14981572 | 0.55 |
|
|
|
| chr8_+_3364453 | 0.55 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr23_+_22858063 | 0.55 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr21_-_25758145 | 0.55 |
|
|
|
| chr2_+_15380054 | 0.55 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr6_+_59580554 | 0.55 |
|
|
|
| chr24_-_34794538 | 0.54 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr9_+_22846082 | 0.54 |
ENSDART00000139270
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 1.3 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.3 | 1.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 | 0.7 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.2 | 1.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.2 | GO:0030237 | female sex determination(GO:0030237) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.6 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.2 | 1.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.1 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 2.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 1.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 1.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.7 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 3.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 0.3 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.2 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.5 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 0.6 | GO:0042373 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.5 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 0.6 | GO:0045980 | negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of purine nucleotide metabolic process(GO:1900543) |
| 0.1 | 1.3 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.3 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.8 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 1.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 2.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.3 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 1.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.8 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.1 | 0.6 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 4.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.3 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 2.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.3 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.8 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 0.4 | GO:1901908 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 0.4 | GO:2000095 | protein localization to ciliary transition zone(GO:1904491) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 1.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.2 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.3 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.5 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.8 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) regulation of cellular response to insulin stimulus(GO:1900076) |
| 0.1 | 0.3 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0035268 | protein mannosylation(GO:0035268) mannosylation(GO:0097502) |
| 0.0 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.7 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.8 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.4 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.7 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 1.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.3 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 1.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.7 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.3 | GO:0051443 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.8 | GO:0009566 | single fertilization(GO:0007338) fertilization(GO:0009566) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.2 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.0 | GO:0098751 | bone cell development(GO:0098751) |
| 0.0 | 0.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0051297 | centrosome organization(GO:0051297) |
| 0.0 | 0.5 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.0 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.9 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.6 | GO:0006865 | amino acid transport(GO:0006865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.4 | 1.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.1 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.2 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.6 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.1 | 3.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 2.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 0.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 1.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 3.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 1.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 1.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 1.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.2 | 0.6 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 1.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.8 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 0.6 | GO:0000403 | Y-form DNA binding(GO:0000403) double-strand/single-strand DNA junction binding(GO:0000406) flap-structured DNA binding(GO:0070336) |
| 0.2 | 1.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.5 | GO:0016844 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.2 | 0.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 1.3 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.4 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.9 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.9 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.8 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.4 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 1.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.5 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 2.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0043878 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.0 | 0.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 2.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.7 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 0.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |