Project
DANIO-CODE
Navigation
Downloads
Results for dlx3b
Z-value: 0.52
Transcription factors associated with dlx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx3b
|
ENSDARG00000014626 | distal-less homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx3b | dr10_dc_chr12_+_5673442_5673506 | -0.23 | 4.0e-01 | Click! |
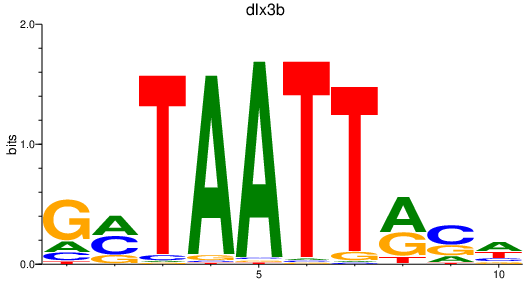
Activity profile of dlx3b motif
Sorted Z-values of dlx3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_11562388 | 0.71 |
ENSDART00000139289
|
cast
|
calpastatin |
| chr14_+_23420053 | 0.71 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| KN150266v1_-_68652 | 0.61 |
|
|
|
| KN150456v1_-_19515 | 0.56 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr21_+_25740782 | 0.54 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr24_-_14446593 | 0.52 |
|
|
|
| chr24_-_23793117 | 0.51 |
|
|
|
| chr19_-_20819477 | 0.44 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr8_-_31098193 | 0.44 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| KN150030v1_-_22613 | 0.43 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr10_-_2944190 | 0.43 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| KN150266v1_-_68620 | 0.42 |
|
|
|
| chr17_+_16038358 | 0.41 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr18_-_43890836 | 0.40 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr5_-_38571198 | 0.40 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr14_+_23419864 | 0.37 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr8_-_23759076 | 0.37 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr19_+_39100820 | 0.34 |
|
|
|
| chr21_-_32747592 | 0.34 |
|
|
|
| chr14_+_23419894 | 0.33 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr23_+_22763739 | 0.31 |
|
|
|
| chr11_-_38856849 | 0.30 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr8_-_31098300 | 0.30 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr19_+_21208954 | 0.29 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr15_+_14656797 | 0.27 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr12_+_20290671 | 0.26 |
ENSDART00000115015
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr5_+_58564645 | 0.26 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr12_+_22459371 | 0.26 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_+_21209328 | 0.25 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr17_+_16038103 | 0.25 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_+_21563703 | 0.25 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr23_+_29017954 | 0.24 |
ENSDART00000140291
|
CR677513.1
|
ENSDARG00000093890 |
| chr6_-_40715613 | 0.23 |
ENSDART00000153702
|
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr16_+_4350448 | 0.22 |
|
|
|
| chr23_+_22763789 | 0.22 |
|
|
|
| chr23_+_17912784 | 0.22 |
ENSDART00000104647
|
prim1
|
primase, DNA, polypeptide 1 |
| chr24_-_14446522 | 0.22 |
|
|
|
| KN150030v1_-_22572 | 0.22 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr21_-_11562321 | 0.21 |
ENSDART00000144370
|
cast
|
calpastatin |
| chr12_+_20290592 | 0.20 |
ENSDART00000115015
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr12_+_22459218 | 0.20 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr16_-_13723295 | 0.20 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr5_+_58564789 | 0.19 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr10_-_2944221 | 0.19 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr23_+_17912834 | 0.19 |
ENSDART00000104647
|
prim1
|
primase, DNA, polypeptide 1 |
| chr4_+_14728603 | 0.18 |
ENSDART00000131666
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr16_-_13723352 | 0.18 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr8_-_21110183 | 0.18 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr24_-_14447825 | 0.17 |
|
|
|
| KN149710v1_+_38638 | 0.17 |
|
|
|
| KN150030v1_-_22642 | 0.17 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr18_+_41242278 | 0.17 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr12_+_20290791 | 0.16 |
ENSDART00000115015
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr19_+_46567459 | 0.15 |
ENSDART00000164055
|
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr15_-_5912105 | 0.15 |
|
|
|
| chr17_-_8413036 | 0.15 |
ENSDART00000154713
|
fzd3b
|
frizzled class receptor 3b |
| chr10_+_6925975 | 0.14 |
|
|
|
| chr5_-_56294194 | 0.14 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_-_42148337 | 0.13 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr2_+_10209233 | 0.13 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr19_+_32000438 | 0.13 |
ENSDART00000150910
|
gmnn
|
geminin, DNA replication inhibitor |
| chr16_-_13723778 | 0.12 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr16_-_4350251 | 0.12 |
|
|
|
| chr8_-_1833095 | 0.12 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr8_-_21110428 | 0.11 |
ENSDART00000135938
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr5_+_26925392 | 0.11 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr7_+_72925910 | 0.11 |
ENSDART00000175267
ENSDART00000179638 |
CABZ01067170.1
|
ENSDARG00000106453 |
| chr24_-_16878970 | 0.11 |
ENSDART00000106058
ENSDART00000106057 |
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr16_+_27837465 | 0.10 |
|
|
|
| chr11_-_6442836 | 0.09 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr24_-_14447136 | 0.09 |
|
|
|
| chr4_+_13587809 | 0.09 |
ENSDART00000138201
|
tnpo3
|
transportin 3 |
| chr19_-_8849482 | 0.09 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr16_-_29259838 | 0.09 |
ENSDART00000178967
|
mef2d
|
myocyte enhancer factor 2d |
| chr20_-_16366248 | 0.08 |
|
|
|
| chr14_-_6901783 | 0.08 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr15_+_14656886 | 0.08 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr4_+_11465367 | 0.08 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr8_+_18552850 | 0.08 |
ENSDART00000177476
|
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr22_-_36816215 | 0.08 |
|
|
|
| chr1_-_57301027 | 0.07 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr8_-_21110262 | 0.07 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr3_-_26052785 | 0.07 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr24_-_14447655 | 0.07 |
|
|
|
| chr6_-_40715579 | 0.07 |
ENSDART00000157113
ENSDART00000154810 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr21_-_11561793 | 0.07 |
ENSDART00000171708
ENSDART00000081614 |
cast
|
calpastatin |
| chr19_+_21783204 | 0.07 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr25_-_3345176 | 0.07 |
ENSDART00000029067
|
hbp1
|
HMG-box transcription factor 1 |
| chr20_+_28901045 | 0.06 |
ENSDART00000153046
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr3_-_16569378 | 0.06 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr16_+_4350419 | 0.06 |
|
|
|
| chr11_-_44539726 | 0.06 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr20_-_22170411 | 0.06 |
ENSDART00000155568
|
BX088688.3
|
ENSDARG00000097598 |
| chr14_-_4117151 | 0.06 |
|
|
|
| chr2_+_40424102 | 0.05 |
ENSDART00000170089
|
epha4b
|
eph receptor A4b |
| chr24_-_16884946 | 0.05 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr7_-_64172974 | 0.05 |
ENSDART00000172619
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr18_+_41242323 | 0.05 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr7_+_16696469 | 0.04 |
|
|
|
| chr11_+_38013238 | 0.04 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr2_-_15656155 | 0.04 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr1_-_10373506 | 0.04 |
|
|
|
| chr9_-_45096523 | 0.03 |
ENSDART00000110697
|
dnajc10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr23_+_22763861 | 0.02 |
|
|
|
| chr15_-_5911817 | 0.02 |
|
|
|
| chr17_+_24803152 | 0.02 |
ENSDART00000112313
|
wdr43
|
WD repeat domain 43 |
| chr1_+_9633924 | 0.02 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr16_+_42567668 | 0.01 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr10_-_34927807 | 0.01 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr7_+_28896401 | 0.01 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr7_-_64173039 | 0.01 |
ENSDART00000172619
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_-_44539778 | 0.01 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr2_-_28687065 | 0.01 |
ENSDART00000164657
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr5_-_31726751 | 0.01 |
|
|
|
| chr7_+_65532871 | 0.00 |
ENSDART00000082740
|
CT573494.2
|
ENSDARG00000097673 |
| chr13_-_36164510 | 0.00 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr6_-_16156294 | 0.00 |
ENSDART00000160809
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.7 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 1.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.4 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |