Project
DANIO-CODE
Navigation
Downloads
Results for dlx4a+dlx4b_dlx2a+dlx2b
Z-value: 0.77
Transcription factors associated with dlx4a+dlx4b_dlx2a+dlx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx4a
|
ENSDARG00000011956 | distal-less homeobox 4a |
|
dlx4b
|
ENSDARG00000071560 | distal-less homeobox 4b |
|
dlx2b
|
ENSDARG00000017174 | distal-less homeobox 2b |
|
dlx2a
|
ENSDARG00000079964 | distal-less homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx4a | dr10_dc_chr3_+_34540552_34540609 | 0.86 | 2.3e-05 | Click! |
| dlx2a | dr10_dc_chr9_-_3429153_3429199 | 0.69 | 2.8e-03 | Click! |
| dlx4b | dr10_dc_chr12_-_5693715_5693760 | 0.69 | 3.2e-03 | Click! |
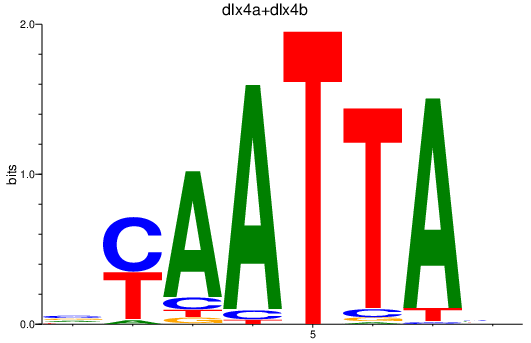
Activity profile of dlx4a+dlx4b_dlx2a+dlx2b motif
Sorted Z-values of dlx4a+dlx4b_dlx2a+dlx2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx4a+dlx4b_dlx2a+dlx2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_24221087 | 3.74 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr2_-_23348612 | 2.50 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr8_+_21321765 | 2.47 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr21_+_28441951 | 2.35 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr20_-_22576513 | 2.00 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_-_35928594 | 1.99 |
|
|
|
| chr20_-_29517770 | 1.93 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr7_+_30516734 | 1.71 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr2_+_49358871 | 1.70 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr16_+_46145286 | 1.52 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr17_-_31642673 | 1.50 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr14_+_20045365 | 1.46 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr20_-_40823307 | 1.46 |
ENSDART00000061261
|
cx43
|
connexin 43 |
| chr16_-_17439735 | 1.45 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr9_-_41608298 | 1.38 |
|
|
|
| chr2_-_36058327 | 1.38 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr16_-_13733759 | 1.37 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr18_-_43890514 | 1.37 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr3_+_17210396 | 1.35 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr23_-_12310778 | 1.33 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr13_+_27654900 | 1.28 |
ENSDART00000160550
|
CR356236.1
|
ENSDARG00000102040 |
| chr16_+_23172295 | 1.27 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr8_-_15091823 | 1.26 |
ENSDART00000142358
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr15_-_20088439 | 1.25 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr16_+_29060022 | 1.24 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr7_-_27877036 | 1.22 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr15_-_1858350 | 1.21 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr2_+_29992879 | 1.19 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr13_-_31304614 | 1.18 |
|
|
|
| chr7_-_25623974 | 1.18 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr22_+_16509286 | 1.17 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr1_-_25600988 | 1.16 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr11_-_6442836 | 1.13 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr19_-_43037791 | 1.13 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr23_+_28804842 | 1.10 |
ENSDART00000047378
|
sst3
|
somatostatin 3 |
| chr16_+_21104310 | 1.09 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr8_-_23591082 | 1.06 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr2_-_17564676 | 1.04 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr9_+_36316158 | 1.01 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr7_+_54410044 | 1.00 |
|
|
|
| chr24_+_40292617 | 1.00 |
|
|
|
| chr17_+_23278879 | 0.97 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr1_-_50215233 | 0.96 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr13_-_44492897 | 0.96 |
|
|
|
| chr16_+_51319421 | 0.93 |
|
|
|
| chr25_+_34069931 | 0.93 |
|
|
|
| chr4_+_11385828 | 0.91 |
|
|
|
| chr24_+_30343687 | 0.91 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr4_+_12616987 | 0.91 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr20_-_9474672 | 0.88 |
ENSDART00000152674
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr20_-_23526954 | 0.88 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr21_-_17259886 | 0.87 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr1_+_38834751 | 0.85 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr2_+_36718808 | 0.85 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr13_-_49695322 | 0.83 |
|
|
|
| chr12_+_20230575 | 0.81 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr20_-_5184885 | 0.81 |
ENSDART00000114851
|
ccdc85cb
|
coiled-coil domain containing 85C, b |
| chr16_+_13928844 | 0.81 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr5_+_38237115 | 0.80 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr4_+_12617165 | 0.80 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr21_-_9863186 | 0.80 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr21_+_26684617 | 0.77 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr17_-_47893203 | 0.77 |
|
|
|
| chr13_+_25319230 | 0.75 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr19_+_22478256 | 0.74 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr4_+_16896821 | 0.72 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr20_-_35343057 | 0.72 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr4_-_15442828 | 0.71 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr17_-_29102320 | 0.71 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr18_+_48426375 | 0.69 |
|
|
|
| chr7_+_42124857 | 0.68 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr9_-_20562293 | 0.68 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr5_+_3965645 | 0.67 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr6_-_14162846 | 0.67 |
ENSDART00000100762
|
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr17_+_24668907 | 0.66 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr15_-_40391882 | 0.66 |
|
|
|
| chr6_+_23787804 | 0.65 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr15_-_23441268 | 0.63 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr16_+_17481157 | 0.62 |
ENSDART00000173448
|
fam131bb
|
family with sequence similarity 131, member Bb |
| chr6_-_54816183 | 0.62 |
ENSDART00000148462
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr24_+_30343717 | 0.62 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr17_-_44133941 | 0.62 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr16_+_13928376 | 0.61 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr1_-_4757890 | 0.60 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr8_+_28240085 | 0.60 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr20_-_20502615 | 0.60 |
|
|
|
| chr23_-_7740845 | 0.59 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr4_+_12613575 | 0.59 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr6_-_6330710 | 0.59 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr6_-_8075384 | 0.58 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr21_+_28921734 | 0.58 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_+_8895437 | 0.57 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr23_-_28286971 | 0.57 |
|
|
|
| chr7_-_52848084 | 0.56 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr20_-_46458839 | 0.54 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr22_+_16282153 | 0.54 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr3_+_47412661 | 0.53 |
ENSDART00000155648
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr21_+_27379622 | 0.53 |
ENSDART00000108763
|
cfb
|
complement factor B |
| chr8_-_46903869 | 0.53 |
ENSDART00000161462
|
acot7
|
acyl-CoA thioesterase 7 |
| chr11_+_37639045 | 0.53 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr19_-_25842461 | 0.52 |
|
|
|
| chr10_-_35293024 | 0.52 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr21_-_11561855 | 0.51 |
ENSDART00000162426
|
cast
|
calpastatin |
| chr17_-_21179064 | 0.47 |
|
|
|
| chr23_+_34037058 | 0.46 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr4_-_22751641 | 0.45 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr19_-_7439294 | 0.45 |
|
|
|
| chr20_+_28958586 | 0.44 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr15_+_4079643 | 0.44 |
|
|
|
| chr20_-_23527004 | 0.44 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr8_+_20592099 | 0.43 |
ENSDART00000160421
|
nfic
|
nuclear factor I/C |
| chr22_-_7371498 | 0.43 |
|
|
|
| KN149726v1_+_1094 | 0.42 |
|
|
|
| chr18_-_7184699 | 0.40 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr12_-_29190576 | 0.40 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr24_-_11103952 | 0.39 |
|
|
|
| chr8_-_38444845 | 0.38 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr19_+_21783429 | 0.38 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr15_-_25582891 | 0.38 |
|
|
|
| chr2_-_7375138 | 0.37 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr22_-_20141589 | 0.37 |
ENSDART00000085913
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr20_-_48255596 | 0.37 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr13_+_24619864 | 0.37 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr3_+_39514598 | 0.36 |
ENSDART00000156038
|
epn2
|
epsin 2 |
| chr24_+_9272045 | 0.36 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr8_+_44719191 | 0.36 |
ENSDART00000163381
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr7_-_25624212 | 0.35 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr2_+_33343287 | 0.35 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr5_-_67233396 | 0.34 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr13_+_25590508 | 0.34 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr22_-_11106940 | 0.34 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr3_-_32205873 | 0.33 |
ENSDART00000156918
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr12_-_26062065 | 0.33 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr13_-_36785295 | 0.32 |
ENSDART00000167154
|
trim9
|
tripartite motif containing 9 |
| chr17_-_23426752 | 0.32 |
ENSDART00000104715
|
pcgf5a
|
polycomb group ring finger 5a |
| chr10_+_3461461 | 0.31 |
|
|
|
| chr11_-_18068313 | 0.30 |
|
|
|
| chr20_-_10133201 | 0.30 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr17_-_21180028 | 0.30 |
ENSDART00000104708
|
abhd12
|
abhydrolase domain containing 12 |
| chr11_-_37730181 | 0.29 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr16_-_20084451 | 0.29 |
ENSDART00000079155
|
hdac9b
|
histone deacetylase 9b |
| chr21_-_21477423 | 0.29 |
ENSDART00000164076
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr23_+_36016450 | 0.29 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr6_+_3120600 | 0.28 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr16_+_22702982 | 0.28 |
ENSDART00000041625
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr19_+_21783111 | 0.27 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr1_+_11290598 | 0.27 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr7_-_25624128 | 0.26 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr14_+_15698814 | 0.25 |
|
|
|
| chr22_-_20141216 | 0.25 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr8_-_19166630 | 0.25 |
|
|
|
| chr1_-_40536800 | 0.24 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr20_-_37587804 | 0.24 |
ENSDART00000166106
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr5_+_36431824 | 0.22 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr16_+_35708856 | 0.22 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr10_-_41478085 | 0.22 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr3_+_14151127 | 0.21 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr19_-_42759951 | 0.21 |
|
|
|
| chr3_-_16569378 | 0.20 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr5_+_26217533 | 0.19 |
|
|
|
| chr11_+_38013238 | 0.18 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr19_-_20203902 | 0.18 |
ENSDART00000172668
|
CR382300.3
|
ENSDARG00000101330 |
| chr11_-_22143133 | 0.17 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr12_+_22137783 | 0.17 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr15_-_16241341 | 0.16 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr24_+_33697332 | 0.16 |
|
|
|
| chr17_-_44133970 | 0.16 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr5_+_19153421 | 0.15 |
|
|
|
| chr21_-_11561978 | 0.14 |
ENSDART00000132699
|
cast
|
calpastatin |
| chr2_+_50873893 | 0.13 |
|
|
|
| chr17_-_29102271 | 0.13 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr1_-_10373506 | 0.13 |
|
|
|
| chr3_-_8608620 | 0.13 |
|
|
|
| chr6_+_50393779 | 0.13 |
ENSDART00000055502
|
ergic3
|
ERGIC and golgi 3 |
| chr8_+_25015374 | 0.12 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr22_-_23518719 | 0.12 |
ENSDART00000161253
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr4_-_13189038 | 0.12 |
ENSDART00000162277
|
grip1
|
glutamate receptor interacting protein 1 |
| chr23_-_294139 | 0.12 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr19_-_42759922 | 0.11 |
|
|
|
| chr8_+_25015325 | 0.11 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr21_+_29040831 | 0.11 |
ENSDART00000166403
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr10_-_34927807 | 0.11 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr21_+_26684728 | 0.10 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr25_+_6607551 | 0.10 |
ENSDART00000176596
|
FQ377991.1
|
ENSDARG00000108452 |
| chr9_-_21649243 | 0.09 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr6_-_23454902 | 0.09 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr3_+_14151244 | 0.09 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr6_+_40924749 | 0.07 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_+_38165187 | 0.07 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr6_-_23455008 | 0.06 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr24_-_39630728 | 0.05 |
ENSDART00000031486
|
lyrm1
|
LYR motif containing 1 |
| chr12_-_34636654 | 0.04 |
|
|
|
| chr3_+_33309011 | 0.04 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr19_+_20213125 | 0.04 |
ENSDART00000167757
|
hoxa1a
|
homeobox A1a |
| chr21_+_40393503 | 0.03 |
ENSDART00000171244
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr3_+_27655753 | 0.03 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr1_-_54087595 | 0.03 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr2_+_31850752 | 0.02 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr4_+_12613877 | 0.02 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr24_-_14447136 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 0.9 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 2.0 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.2 | 1.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 2.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 0.8 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 1.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 1.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.6 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.1 | 0.9 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.7 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.2 | GO:0050886 | phasic smooth muscle contraction(GO:0014821) endocrine process(GO:0050886) |
| 0.1 | 1.0 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.2 | GO:1902024 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.8 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0061626 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.0 | 0.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 1.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 3.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 1.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.2 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.6 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 1.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 1.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.3 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.7 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 3.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.8 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.6 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 2.5 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.4 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 3.0 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.3 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 2.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |