Project
DANIO-CODE
Navigation
Downloads
Results for dlx5a
Z-value: 0.45
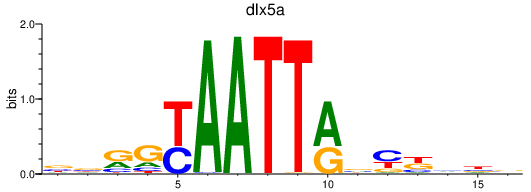
Transcription factors associated with dlx5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dlx5a
|
ENSDARG00000042296 | distal-less homeobox 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dlx5a | dr10_dc_chr19_-_41887040_41887127 | 0.04 | 8.9e-01 | Click! |
Activity profile of dlx5a motif
Sorted Z-values of dlx5a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dlx5a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_12310778 | 0.93 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr2_-_23348612 | 0.78 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr7_-_25623974 | 0.75 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr16_+_14981032 | 0.72 |
|
|
|
| chr7_+_24770873 | 0.66 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr5_+_56885728 | 0.53 |
|
|
|
| chr5_+_68410884 | 0.51 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr7_-_32888309 | 0.49 |
ENSDART00000173461
|
BX571811.2
|
ENSDARG00000105655 |
| chr15_-_18110169 | 0.48 |
|
|
|
| KN149726v1_+_1094 | 0.45 |
|
|
|
| chr7_-_25624212 | 0.45 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr17_+_1986151 | 0.43 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr7_-_25624128 | 0.36 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr16_+_29060022 | 0.34 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr19_+_19939410 | 0.30 |
|
|
|
| chr3_+_13692658 | 0.28 |
ENSDART00000162317
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr3_+_32700750 | 0.26 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr24_-_36144354 | 0.25 |
|
|
|
| chr7_+_56401839 | 0.24 |
ENSDART00000004964
|
nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr15_-_5911817 | 0.24 |
|
|
|
| chr24_-_14447773 | 0.24 |
|
|
|
| chr3_-_23513293 | 0.23 |
ENSDART00000140264
|
eve1
|
even-skipped-like1 |
| chr1_-_39622531 | 0.21 |
|
|
|
| KN150079v1_-_26372 | 0.21 |
ENSDART00000178246
|
CABZ01118121.2
|
ENSDARG00000108979 |
| chr23_+_31986806 | 0.20 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr8_+_456029 | 0.20 |
ENSDART00000051776
|
srp19
|
signal recognition particle 19 |
| chr17_+_24803152 | 0.19 |
ENSDART00000112313
|
wdr43
|
WD repeat domain 43 |
| chr3_+_25888520 | 0.18 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr14_+_10984191 | 0.18 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| chr24_-_36143918 | 0.18 |
|
|
|
| chr19_+_15617624 | 0.16 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr8_-_455930 | 0.16 |
ENSDART00000051777
|
zgc:101664
|
zgc:101664 |
| chr19_-_28699617 | 0.16 |
ENSDART00000151756
|
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr20_-_2936289 | 0.15 |
ENSDART00000104667
|
cdk19
|
cyclin-dependent kinase 19 |
| chr16_+_35708856 | 0.14 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr11_-_37452764 | 0.14 |
ENSDART00000023388
|
yod1
|
YOD1 deubiquitinase |
| chr6_-_21733038 | 0.14 |
|
|
|
| chr16_+_17808801 | 0.13 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr3_+_17387551 | 0.12 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| KN149830v1_-_23361 | 0.12 |
|
|
|
| chr17_+_37306052 | 0.11 |
ENSDART00000134000
|
tmem62
|
transmembrane protein 62 |
| chr23_+_34037058 | 0.10 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr5_-_56885580 | 0.10 |
|
|
|
| chr24_-_34794463 | 0.09 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr25_-_18855316 | 0.09 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr13_-_4095406 | 0.08 |
ENSDART00000113060
ENSDART00000138892 |
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr2_-_30071815 | 0.08 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr1_-_13302745 | 0.08 |
ENSDART00000011414
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr24_+_16402613 | 0.07 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr24_-_14447519 | 0.06 |
|
|
|
| chr10_-_35205597 | 0.06 |
ENSDART00000131291
|
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr3_+_32700882 | 0.05 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr20_-_2991247 | 0.05 |
ENSDART00000160701
|
ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr8_-_44247277 | 0.05 |
|
|
|
| chr8_-_52426509 | 0.04 |
ENSDART00000015081
|
ENSDARG00000011208
|
ENSDARG00000011208 |
| chr21_+_7737034 | 0.04 |
|
|
|
| chr1_-_10373506 | 0.03 |
|
|
|
| chr19_+_21208954 | 0.03 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr24_-_14446522 | 0.03 |
|
|
|
| chr19_+_5562075 | 0.02 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr23_-_31986482 | 0.01 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr9_-_12604260 | 0.01 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr9_+_13258378 | 0.01 |
ENSDART00000141705
ENSDART00000154879 |
carf
|
calcium responsive transcription factor |
| chr17_-_117828 | 0.01 |
|
|
|
| chr6_-_8075384 | 0.00 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr14_+_10983831 | 0.00 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| KN150642v1_+_9419 | 0.00 |
ENSDART00000159069
|
atoh1b
|
atonal bHLH transcription factor 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002821 | regulation of lymphocyte mediated immunity(GO:0002706) positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) regulation of B cell activation(GO:0050864) |
| 0.0 | 0.1 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0030431 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |