Project
DANIO-CODE
Navigation
Downloads
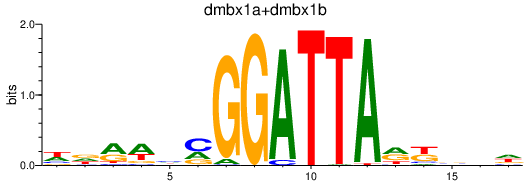
Results for dmbx1a+dmbx1b
Z-value: 0.56
Transcription factors associated with dmbx1a+dmbx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
dmbx1b
|
ENSDARG00000002510 | diencephalon/mesencephalon homeobox 1b |
|
dmbx1a
|
ENSDARG00000009922 | diencephalon/mesencephalon homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| dmbx1b | dr10_dc_chr6_-_3413472_3413503 | -0.18 | 5.0e-01 | Click! |
| dmbx1a | dr10_dc_chr2_-_10394639_10394685 | -0.14 | 6.0e-01 | Click! |
Activity profile of dmbx1a+dmbx1b motif
Sorted Z-values of dmbx1a+dmbx1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of dmbx1a+dmbx1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_37732100 | 2.10 |
|
|
|
| chr11_-_34367105 | 0.87 |
ENSDART00000157625
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr10_+_8670564 | 0.83 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr1_-_11439671 | 0.78 |
ENSDART00000164817
|
mtp
|
microsomal triglyceride transfer protein |
| chr4_-_16639774 | 0.76 |
ENSDART00000125323
|
caprin2
|
caprin family member 2 |
| chr15_-_41777233 | 0.68 |
ENSDART00000154230
|
ftr90
|
finTRIM family, member 90 |
| chr25_-_34616997 | 0.58 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr23_-_3816492 | 0.54 |
ENSDART00000136394
|
hmga1a
|
high mobility group AT-hook 1a |
| chr8_+_46319434 | 0.48 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr5_+_3597060 | 0.46 |
|
|
|
| chr3_+_22962233 | 0.46 |
ENSDART00000142884
|
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr23_-_3816422 | 0.45 |
ENSDART00000132205
|
hmga1a
|
high mobility group AT-hook 1a |
| chr18_+_5616001 | 0.42 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr1_+_35217331 | 0.41 |
|
|
|
| chr12_+_33793468 | 0.40 |
ENSDART00000130853
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr15_-_6979997 | 0.37 |
ENSDART00000169944
|
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr22_+_13862110 | 0.36 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr20_-_21773202 | 0.36 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_-_22099244 | 0.36 |
ENSDART00000161298
|
nono
|
non-POU domain containing, octamer-binding |
| chr12_+_33793663 | 0.36 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr15_-_36072087 | 0.33 |
ENSDART00000076229
|
irs1
|
insulin receptor substrate 1 |
| chr20_-_21773327 | 0.32 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_-_22098977 | 0.31 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr10_-_8029671 | 0.30 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr13_-_31807963 | 0.28 |
ENSDART00000026726
|
diexf
|
digestive organ expansion factor homolog |
| chr18_-_10744619 | 0.26 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr15_-_7340623 | 0.26 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
solute carrier family 7 member 1 |
| chr16_+_25096883 | 0.25 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr11_-_2437361 | 0.24 |
|
|
|
| chr5_-_68408107 | 0.23 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr22_-_36717838 | 0.22 |
|
|
|
| chr18_+_50528517 | 0.22 |
ENSDART00000151647
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| KN149926v1_+_2353 | 0.21 |
|
|
|
| chr5_-_4750799 | 0.21 |
ENSDART00000130576
|
mvb12ba
|
multivesicular body subunit 12Ba |
| chr19_+_24741199 | 0.20 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr25_-_34617182 | 0.20 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr16_+_25096821 | 0.19 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr12_+_33793495 | 0.18 |
ENSDART00000152988
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr6_-_39655998 | 0.17 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr2_-_38380883 | 0.17 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr15_-_25825347 | 0.17 |
ENSDART00000155332
ENSDART00000154029 |
CT027805.1
|
ENSDARG00000097316 |
| chr5_-_23525132 | 0.17 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr15_-_5575675 | 0.17 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr14_+_30390987 | 0.17 |
|
|
|
| chr6_-_39656043 | 0.17 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr2_+_45119713 | 0.16 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr13_-_38666198 | 0.16 |
ENSDART00000162858
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr23_-_29627060 | 0.16 |
ENSDART00000166554
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr5_-_8313115 | 0.16 |
ENSDART00000170921
|
fgf10b
|
fibroblast growth factor 10b |
| chr5_-_68408171 | 0.15 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr25_-_34617243 | 0.15 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr23_-_27123338 | 0.14 |
ENSDART00000142324
ENSDART00000133249 |
ENSDARG00000025766
|
ENSDARG00000025766 |
| chr5_-_25133229 | 0.13 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr9_+_31265296 | 0.13 |
ENSDART00000132478
|
BX470183.1
|
ENSDARG00000095476 |
| chr5_-_4750869 | 0.13 |
ENSDART00000164377
|
mvb12ba
|
multivesicular body subunit 12Ba |
| chr2_-_38380637 | 0.13 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr23_-_533016 | 0.13 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr21_-_13599764 | 0.12 |
|
|
|
| chr18_+_5066229 | 0.12 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr19_+_24741351 | 0.12 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr25_-_31970637 | 0.11 |
ENSDART00000006124
|
cops2
|
COP9 signalosome subunit 2 |
| chr23_-_532974 | 0.11 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr13_+_40689102 | 0.11 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr25_-_34617301 | 0.11 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr15_-_8215797 | 0.10 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr4_-_4247251 | 0.09 |
ENSDART00000150279
|
cd9b
|
CD9 molecule b |
| chr25_-_34616837 | 0.09 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr13_-_36456275 | 0.09 |
ENSDART00000143682
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr5_-_25133456 | 0.08 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr2_+_30800532 | 0.08 |
|
|
|
| chr15_-_5575746 | 0.08 |
ENSDART00000130297
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr4_-_74907755 | 0.07 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr12_+_31614163 | 0.07 |
ENSDART00000152973
|
RNF157
|
ring finger protein 157 |
| chr13_-_36456350 | 0.07 |
ENSDART00000143682
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr17_+_24668907 | 0.06 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr17_+_10437343 | 0.06 |
ENSDART00000167188
|
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr14_-_49951804 | 0.04 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr19_-_1571878 | 0.04 |
|
|
|
| chr18_-_11216036 | 0.04 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr18_+_5616306 | 0.03 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr2_-_9946238 | 0.03 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr8_-_40412363 | 0.03 |
ENSDART00000161142
|
tbc1d10aa
|
TBC1 domain family, member 10Aa |
| KN149926v1_+_1959 | 0.03 |
|
|
|
| chr3_+_47412661 | 0.03 |
ENSDART00000155648
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr21_-_3619164 | 0.02 |
ENSDART00000133980
|
scamp1
|
secretory carrier membrane protein 1 |
| chr2_-_9946148 | 0.02 |
ENSDART00000175460
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr25_+_35861851 | 0.01 |
|
|
|
| chr17_-_8498935 | 0.01 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr18_+_5616238 | 0.00 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.2 | 0.8 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.2 | 0.5 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 0.3 | GO:0044803 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) purine deoxyribonucleotide catabolic process(GO:0009155) purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.2 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.5 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.2 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.2 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |