Project
DANIO-CODE
Navigation
Downloads
Results for e2f3
Z-value: 0.79
Transcription factors associated with e2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f3
|
ENSDARG00000070463 | E2F transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f3 | dr10_dc_chr19_-_29707065_29707294 | 0.92 | 4.2e-07 | Click! |
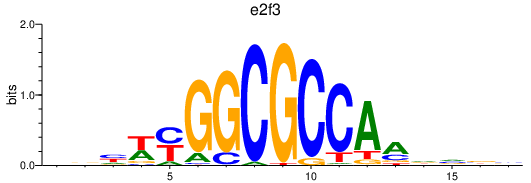
Activity profile of e2f3 motif
Sorted Z-values of e2f3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29219700 | 2.54 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr14_-_30578373 | 1.91 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr8_-_3354396 | 1.76 |
ENSDART00000169430
ENSDART00000167187 ENSDART00000170478 |
FUT9 (1 of many)
fut9b
|
fucosyltransferase 9 fucosyltransferase 9b |
| chr11_-_11809024 | 1.65 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr16_-_37518912 | 1.64 |
|
|
|
| chr14_+_14536088 | 1.52 |
ENSDART00000158291
|
slbp
|
stem-loop binding protein |
| chr20_+_27006101 | 1.44 |
ENSDART00000153213
|
rwdd2b
|
RWD domain containing 2B |
| chr19_-_44161905 | 1.35 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr9_-_7235236 | 1.33 |
ENSDART00000092435
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr10_+_16543480 | 1.31 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr7_-_33965017 | 1.28 |
|
|
|
| chr20_+_26495546 | 1.22 |
ENSDART00000036942
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr23_+_37476457 | 1.19 |
ENSDART00000178064
|
AL954146.3
|
ENSDARG00000108629 |
| chr7_+_33965084 | 1.15 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr3_-_54352535 | 1.13 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr9_+_22866612 | 1.04 |
ENSDART00000131429
ENSDART00000080005 |
itgb5
|
integrin, beta 5 |
| chr25_+_35790000 | 1.02 |
ENSDART00000152195
|
si:ch211-113a14.18
|
si:ch211-113a14.18 |
| chr19_-_44161951 | 0.99 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr14_-_21779536 | 0.97 |
|
|
|
| chr6_-_32001100 | 0.93 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr17_+_6378694 | 0.93 |
ENSDART00000062952
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr6_+_60117223 | 0.82 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr17_-_31061338 | 0.79 |
|
|
|
| chr9_-_7235272 | 0.78 |
ENSDART00000174720
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr5_+_12894381 | 0.75 |
ENSDART00000051669
|
tctn2
|
tectonic family member 2 |
| chr9_-_2886135 | 0.67 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr19_-_10295398 | 0.63 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr21_-_21477462 | 0.61 |
ENSDART00000031205
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr11_-_11808973 | 0.57 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr11_+_44738699 | 0.56 |
ENSDART00000172301
ENSDART00000170118 |
tk1
|
thymidine kinase 1, soluble |
| chr16_-_12362566 | 0.55 |
ENSDART00000116542
|
U7
|
U7 small nuclear RNA |
| chr20_+_26495186 | 0.54 |
ENSDART00000078093
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr17_+_6378932 | 0.50 |
ENSDART00000062952
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr6_-_37534996 | 0.40 |
ENSDART00000147826
|
jade3
|
jade family PHD finger 3 |
| chr14_+_21779325 | 0.40 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr6_-_49323847 | 0.40 |
ENSDART00000155232
|
BX569793.1
|
ENSDARG00000096832 |
| chr20_+_27006042 | 0.40 |
ENSDART00000153149
ENSDART00000138249 |
rwdd2b
|
RWD domain containing 2B |
| chr7_+_1372988 | 0.27 |
|
|
|
| KN150648v1_+_877 | 0.22 |
|
|
|
| chr19_-_10295122 | 0.15 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr2_+_21431575 | 0.14 |
ENSDART00000109568
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr17_-_32826829 | 0.14 |
ENSDART00000077459
|
smyd2a
|
SET and MYND domain containing 2a |
| chr8_-_11950254 | 0.08 |
ENSDART00000005140
|
med27
|
mediator complex subunit 27 |
| chr20_+_26495276 | 0.08 |
ENSDART00000078093
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr1_+_9202797 | 0.05 |
ENSDART00000135702
|
rgs11
|
regulator of G protein signaling 11 |
| chr1_+_35990022 | 0.04 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr5_-_6384926 | 0.02 |
ENSDART00000099977
|
mex3c
|
mex-3 RNA binding family member C |
| chr2_-_44491143 | 0.01 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr10_+_16543525 | 0.01 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 2.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 1.1 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.2 | 1.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 2.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.9 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 1.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.3 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 2.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.7 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.6 | GO:0006213 | pyrimidine nucleoside metabolic process(GO:0006213) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.3 | GO:0005764 | lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 1.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |