Project
DANIO-CODE
Navigation
Downloads
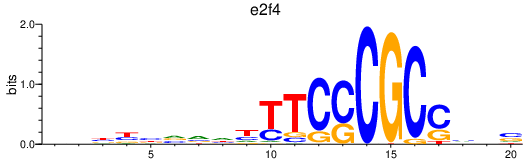
Results for e2f4
Z-value: 0.77
Transcription factors associated with e2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f4
|
ENSDARG00000101578 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f4 | dr10_dc_chr25_+_17308788_17308907 | -0.24 | 3.8e-01 | Click! |
Activity profile of e2f4 motif
Sorted Z-values of e2f4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_17289857 | 1.41 |
ENSDART00000178686
|
lrmp
|
lymphoid-restricted membrane protein |
| chr12_+_47448318 | 1.39 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr10_+_6925373 | 1.28 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr25_-_34619603 | 1.11 |
ENSDART00000114767
|
FQ312024.1
|
ENSDARG00000076129 |
| chr7_-_6209659 | 1.10 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr25_-_35820568 | 1.09 |
ENSDART00000152766
|
CR354435.4
|
Histone H2B 1/2 |
| chr7_+_5839103 | 1.07 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr25_+_34562216 | 0.99 |
ENSDART00000154655
|
CU302436.4
|
ENSDARG00000092743 |
| chr7_-_73631286 | 0.99 |
ENSDART00000129254
|
zgc:173552
|
zgc:173552 |
| chr20_-_28531087 | 0.93 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr19_-_48731766 | 0.87 |
ENSDART00000170726
|
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr7_+_24610446 | 0.86 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr14_-_27040866 | 0.85 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr25_+_34553079 | 0.84 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr7_+_24543671 | 0.79 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr25_+_22222336 | 0.77 |
ENSDART00000154065
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr23_-_19905270 | 0.75 |
ENSDART00000054479
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr20_-_54160322 | 0.75 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr11_-_27378184 | 0.69 |
ENSDART00000157337
|
CR931782.1
|
ENSDARG00000097455 |
| chr20_-_54160297 | 0.66 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr25_-_34604544 | 0.66 |
ENSDART00000154856
|
CU302436.5
|
Histone H3.2 |
| chr5_+_19944746 | 0.61 |
ENSDART00000124545
|
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr11_+_31417477 | 0.60 |
ENSDART00000139900
|
diaph3
|
diaphanous-related formin 3 |
| chr7_+_73594790 | 0.59 |
ENSDART00000123081
|
zgc:173552
|
zgc:173552 |
| chr13_+_15802079 | 0.58 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr5_-_22466262 | 0.57 |
ENSDART00000172549
|
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr20_+_34091702 | 0.57 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr7_+_6402162 | 0.56 |
|
|
|
| chr3_-_8011385 | 0.54 |
ENSDART00000169757
|
ubn2b
|
ubinuclein 2b |
| chr3_+_21928413 | 0.54 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr13_-_22776767 | 0.53 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr20_-_28531019 | 0.52 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr13_+_15802118 | 0.50 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr14_-_16469435 | 0.50 |
ENSDART00000113711
|
zgc:123335
|
zgc:123335 |
| chr4_-_16611960 | 0.49 |
|
|
|
| chr7_+_6402122 | 0.48 |
|
|
|
| chr25_-_12728011 | 0.47 |
ENSDART00000169126
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr6_+_50382153 | 0.46 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr3_+_62051674 | 0.45 |
ENSDART00000113144
|
zgc:173575
|
zgc:173575 |
| chr7_-_6293116 | 0.42 |
ENSDART00000173413
|
zgc:112234
|
zgc:112234 |
| chr7_-_6323808 | 0.42 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr11_+_24110796 | 0.41 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr22_-_26254136 | 0.41 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr21_+_30684976 | 0.41 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr8_+_8908955 | 0.40 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr4_-_27109874 | 0.40 |
ENSDART00000146459
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr14_-_16469491 | 0.40 |
ENSDART00000113711
|
zgc:123335
|
zgc:123335 |
| chr3_+_62101056 | 0.39 |
|
|
|
| chr19_+_48731885 | 0.39 |
|
|
|
| chr13_-_22776819 | 0.38 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr19_+_14490203 | 0.37 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr16_+_54906490 | 0.34 |
ENSDART00000126646
|
ENSDARG00000091079
|
ENSDARG00000091079 |
| KN149926v1_+_2353 | 0.33 |
|
|
|
| chr3_+_62051743 | 0.31 |
ENSDART00000168063
|
zgc:173575
|
zgc:173575 |
| chr7_-_6323664 | 0.30 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr18_+_44635321 | 0.30 |
|
|
|
| chr25_+_181958 | 0.28 |
ENSDART00000155412
|
RPS17
|
ribosomal protein S17 |
| chr8_-_43733891 | 0.26 |
ENSDART00000166581
|
ep400
|
E1A binding protein p400 |
| chr25_+_34553195 | 0.26 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr20_+_27193945 | 0.26 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr4_-_16611908 | 0.26 |
|
|
|
| chr18_+_17032264 | 0.25 |
ENSDART00000100160
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr18_+_39345919 | 0.24 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr23_-_20592068 | 0.24 |
ENSDART00000153767
|
CR352228.1
|
ENSDARG00000097895 |
| chr18_-_40763107 | 0.22 |
ENSDART00000130397
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| KN149926v1_+_1959 | 0.20 |
|
|
|
| chr17_-_11214364 | 0.20 |
ENSDART00000155976
|
FP102895.1
|
ENSDARG00000097935 |
| chr23_+_20592121 | 0.20 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr22_-_26254217 | 0.18 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr3_-_23473646 | 0.18 |
ENSDART00000078423
|
atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr5_+_38499102 | 0.18 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr2_-_57973733 | 0.16 |
ENSDART00000174670
ENSDART00000176252 |
CABZ01093382.1
|
ENSDARG00000105978 |
| chr7_-_6280179 | 0.15 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr25_+_34557335 | 0.14 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr14_-_27041347 | 0.13 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr17_+_21495087 | 0.10 |
ENSDART00000154633
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr6_+_54798741 | 0.09 |
|
|
|
| chr21_-_27376521 | 0.09 |
ENSDART00000131646
|
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr13_-_45805308 | 0.08 |
ENSDART00000166442
|
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr14_-_6096120 | 0.07 |
|
|
|
| chr7_-_6293333 | 0.06 |
ENSDART00000173413
|
zgc:112234
|
zgc:112234 |
| chr24_+_17201072 | 0.05 |
ENSDART00000024722
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr16_-_32174276 | 0.03 |
ENSDART00000137699
|
cnot3a
|
CCR4-NOT transcription complex, subunit 3a |
| chr2_+_37854303 | 0.01 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr2_-_37854616 | 0.00 |
ENSDART00000171409
|
mettl17
|
methyltransferase like 17 |
| chr21_-_27376494 | 0.00 |
ENSDART00000011551
ENSDART00000148161 |
slc29a2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.3 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 1.3 | GO:0048608 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.1 | 1.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.2 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 1.4 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 1.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.8 | GO:0051225 | centrosome cycle(GO:0007098) spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 1.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 1.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0009055 | electron carrier activity(GO:0009055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |