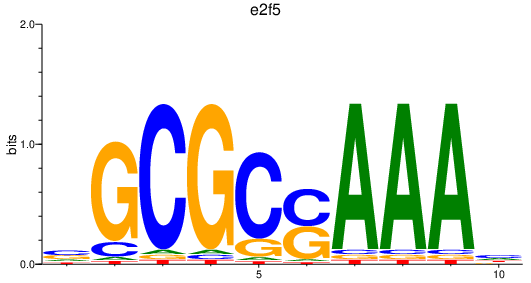
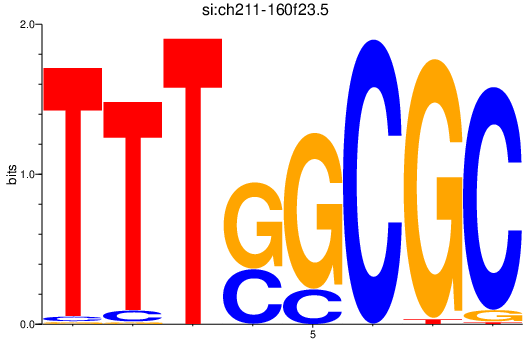
Project
DANIO-CODE
Navigation
Downloads
Results for e2f5_si:ch211-160f23.5
Z-value: 2.15
Transcription factors associated with e2f5_si:ch211-160f23.5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f5
|
ENSDARG00000038812 | E2F transcription factor 5 |
|
si_ch211-160f23.5
|
ENSDARG00000079233 | si_ch211-160f23.5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f5 | dr10_dc_chr2_-_31703111_31703237 | 0.71 | 2.2e-03 | Click! |
| E2F2 | dr10_dc_chr17_-_27247516_27247601 | -0.37 | 1.6e-01 | Click! |
Activity profile of e2f5_si:ch211-160f23.5 motif
Sorted Z-values of e2f5_si:ch211-160f23.5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f5_si:ch211-160f23.5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22222388 | 8.58 |
ENSDART00000154376
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr15_-_17163526 | 8.47 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr2_-_29939721 | 8.18 |
ENSDART00000031130
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr11_-_11809024 | 7.67 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr3_-_54352535 | 6.84 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr19_-_6925090 | 6.31 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr1_-_7869105 | 5.60 |
ENSDART00000147763
|
zgc:112980
|
zgc:112980 |
| chr6_+_50452131 | 4.96 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr25_+_29219700 | 4.79 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr7_+_55217087 | 4.71 |
ENSDART00000149915
|
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr20_+_35535503 | 4.50 |
ENSDART00000153249
|
tdrd6
|
tudor domain containing 6 |
| chr19_+_39689450 | 4.27 |
|
|
|
| chr23_-_36207066 | 4.03 |
ENSDART00000147598
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr8_-_25741895 | 3.92 |
ENSDART00000140451
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr2_+_44659334 | 3.83 |
ENSDART00000155017
|
pask
|
PAS domain containing serine/threonine kinase |
| chr5_-_12242927 | 3.65 |
ENSDART00000137705
|
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr20_+_39441958 | 3.62 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr16_+_41065840 | 3.58 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr9_-_2886135 | 3.53 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr7_-_64745132 | 3.50 |
ENSDART00000112166
|
fam60al
|
family with sequence similarity 60, member A, like |
| chr14_+_21831841 | 3.41 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr3_+_25868700 | 3.19 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr25_+_22222336 | 3.19 |
ENSDART00000154065
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_-_33965017 | 3.14 |
|
|
|
| chr5_+_37019043 | 3.11 |
|
|
|
| chr6_-_1685041 | 3.10 |
|
|
|
| chr2_+_37855019 | 3.09 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr7_+_33965084 | 3.09 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr13_+_7923056 | 3.04 |
ENSDART00000080465
|
hells
|
helicase, lymphoid-specific |
| chr19_-_43359719 | 3.02 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr19_-_29707065 | 3.00 |
ENSDART00000130815
ENSDART00000172590 ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr16_-_37518912 | 2.96 |
|
|
|
| chr15_+_17163165 | 2.94 |
|
|
|
| chr10_+_21477579 | 2.94 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr11_-_11808973 | 2.89 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr5_+_37018446 | 2.88 |
|
|
|
| chr23_+_45486958 | 2.88 |
ENSDART00000111126
|
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr19_-_20819477 | 2.85 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr3_+_26113651 | 2.82 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr7_+_41532360 | 2.71 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr19_-_20819101 | 2.70 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr8_-_22639164 | 2.65 |
|
|
|
| chr22_-_38321005 | 2.62 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_+_26113393 | 2.50 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr6_+_33947023 | 2.36 |
ENSDART00000157397
|
orc1
|
origin recognition complex, subunit 1 |
| chr2_-_40857927 | 2.31 |
ENSDART00000157099
|
CT583650.1
|
ENSDARG00000096869 |
| chr18_+_15303434 | 2.30 |
ENSDART00000099777
ENSDART00000170246 |
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr3_-_26113336 | 2.29 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr6_+_50452229 | 2.27 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr8_-_4704361 | 2.26 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr23_-_36349563 | 2.26 |
|
|
|
| chr17_-_33754959 | 2.25 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr3_-_26112995 | 2.20 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr14_-_31375046 | 2.15 |
ENSDART00000173274
|
map7d3
|
MAP7 domain containing 3 |
| chr7_-_41532268 | 2.14 |
ENSDART00000016105
ENSDART00000138800 |
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr8_-_1154220 | 2.12 |
ENSDART00000149969
ENSDART00000016800 |
znf367
|
zinc finger protein 367 |
| chr7_-_68962286 | 2.12 |
|
|
|
| chr19_-_2928582 | 2.09 |
ENSDART00000109130
|
recql4
|
RecQ helicase-like 4 |
| chr7_-_37624329 | 2.09 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr3_-_15960491 | 2.07 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr20_-_44678938 | 2.06 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr18_-_12889296 | 2.05 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr3_+_22204475 | 2.03 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr13_-_12898069 | 2.01 |
ENSDART00000009499
|
whsc1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr7_+_71293928 | 2.01 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr7_+_1443102 | 2.01 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr5_+_69178270 | 2.00 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr20_-_33584483 | 1.96 |
ENSDART00000177645
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr2_+_38073166 | 1.94 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr18_+_50920737 | 1.93 |
ENSDART00000174305
|
cttn
|
cortactin |
| chr21_-_19879125 | 1.92 |
ENSDART00000164317
|
eri1
|
exoribonuclease 1 |
| chr5_+_37018996 | 1.92 |
|
|
|
| chr13_-_35782121 | 1.90 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr19_+_9536231 | 1.89 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr12_+_47448318 | 1.89 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr25_+_29752983 | 1.86 |
ENSDART00000141167
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr19_-_10295398 | 1.86 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr7_-_37624410 | 1.86 |
ENSDART00000084282
|
papd5
|
PAP associated domain containing 5 |
| chr17_-_22552999 | 1.85 |
ENSDART00000141523
|
exo1
|
exonuclease 1 |
| chr6_-_25081182 | 1.85 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 family, member Db |
| chr24_-_35673629 | 1.83 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr18_-_12889369 | 1.81 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr5_+_19944746 | 1.79 |
ENSDART00000124545
|
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr3_+_26212025 | 1.78 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr22_+_2524697 | 1.78 |
|
|
|
| chr19_-_29706969 | 1.78 |
ENSDART00000130815
ENSDART00000172590 ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr21_+_22521573 | 1.77 |
ENSDART00000157593
ENSDART00000167599 |
chek1
|
checkpoint kinase 1 |
| chr5_-_53748764 | 1.75 |
ENSDART00000160781
ENSDART00000160965 ENSDART00000161692 |
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr22_-_10562118 | 1.74 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr5_+_33949157 | 1.74 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr11_+_41777220 | 1.71 |
ENSDART00000172846
|
camta1
|
calmodulin binding transcription activator 1 |
| chr10_-_21404605 | 1.69 |
ENSDART00000125167
|
avd
|
avidin |
| chr19_-_9044008 | 1.64 |
ENSDART00000104657
|
mrps21
|
mitochondrial ribosomal protein S21 |
| chr7_-_40386766 | 1.63 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr8_-_3354396 | 1.63 |
ENSDART00000169430
ENSDART00000167187 ENSDART00000170478 |
FUT9 (1 of many)
fut9b
|
fucosyltransferase 9 fucosyltransferase 9b |
| chr8_-_40287530 | 1.62 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr13_+_20393886 | 1.60 |
ENSDART00000081385
|
ccdc172
|
coiled-coil domain containing 172 |
| chr13_-_12898263 | 1.59 |
ENSDART00000009499
|
whsc1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr5_+_33949689 | 1.58 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr14_-_5103890 | 1.56 |
|
|
|
| chr5_+_58009274 | 1.54 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr22_+_1633211 | 1.54 |
ENSDART00000160854
|
BX649416.3
|
ENSDARG00000104479 |
| chr3_+_36970806 | 1.54 |
ENSDART00000055228
|
psmc3ip
|
PSMC3 interacting protein |
| chr20_+_21369047 | 1.54 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr3_+_22204569 | 1.54 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr8_-_40287837 | 1.52 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr23_-_36207526 | 1.52 |
ENSDART00000142760
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr25_+_26458700 | 1.50 |
|
|
|
| chr1_+_13696375 | 1.48 |
ENSDART00000103346
|
fip1l1b
|
FIP1 like 1b (S. cerevisiae) |
| chr23_-_43396787 | 1.46 |
|
|
|
| chr7_+_23752492 | 1.46 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr9_+_22970052 | 1.45 |
ENSDART00000143972
|
rif1
|
replication timing regulatory factor 1 |
| chr16_+_41065107 | 1.44 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr7_+_71345049 | 1.43 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr24_+_7295201 | 1.43 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr10_+_21477307 | 1.42 |
ENSDART00000064558
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr22_-_1965842 | 1.41 |
ENSDART00000164037
|
FO704584.2
|
ENSDARG00000103579 |
| chr7_+_13275370 | 1.39 |
ENSDART00000020542
|
plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr6_-_34854950 | 1.39 |
ENSDART00000169605
|
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr17_-_26519582 | 1.38 |
ENSDART00000155692
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr2_-_11335644 | 1.38 |
ENSDART00000135450
|
cryz
|
crystallin, zeta (quinone reductase) |
| chr25_+_26457948 | 1.37 |
ENSDART00000153839
|
znf800a
|
zinc finger protein 800a |
| chr7_-_6328372 | 1.37 |
ENSDART00000130760
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr2_-_57826189 | 1.37 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr7_+_31103149 | 1.35 |
ENSDART00000085505
|
ctdspl2b
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2b |
| chr2_-_44924505 | 1.35 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr5_+_33949469 | 1.35 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr23_-_36207591 | 1.35 |
ENSDART00000143340
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr15_+_44160948 | 1.34 |
ENSDART00000110060
|
zgc:165514
|
zgc:165514 |
| chr6_-_37534996 | 1.34 |
ENSDART00000147826
|
jade3
|
jade family PHD finger 3 |
| chr20_-_33584973 | 1.31 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr23_+_40517744 | 1.30 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr20_-_9212139 | 1.30 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr9_-_7235236 | 1.28 |
ENSDART00000092435
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_+_37854303 | 1.26 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr15_-_17163104 | 1.25 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr22_-_3986398 | 1.25 |
ENSDART00000169317
ENSDART00000171262 |
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr5_+_26537518 | 1.25 |
|
|
|
| chr20_-_33584657 | 1.25 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr17_-_26519469 | 1.22 |
ENSDART00000155692
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr23_+_37476457 | 1.21 |
ENSDART00000178064
|
AL954146.3
|
ENSDARG00000108629 |
| chr19_-_2651862 | 1.18 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr23_-_36206989 | 1.18 |
ENSDART00000147598
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr22_-_3134699 | 1.16 |
ENSDART00000159580
|
lmnb2
|
lamin B2 |
| chr8_-_25741931 | 1.16 |
ENSDART00000140451
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr8_+_20383989 | 1.16 |
ENSDART00000009081
|
mob3a
|
MOB kinase activator 3A |
| chr17_-_26519322 | 1.15 |
ENSDART00000122366
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr5_-_22466262 | 1.14 |
ENSDART00000172549
|
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr21_-_30750638 | 1.14 |
ENSDART00000012091
|
rrm1
|
ribonucleotide reductase M1 polypeptide |
| chr7_-_37648061 | 1.14 |
ENSDART00000052368
|
heatr3
|
HEAT repeat containing 3 |
| chr25_-_13307171 | 1.14 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr22_-_8144599 | 1.13 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr23_+_24672673 | 1.11 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr6_+_33946990 | 1.11 |
ENSDART00000157397
|
orc1
|
origin recognition complex, subunit 1 |
| chr22_-_8144696 | 1.10 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr16_+_25693740 | 1.10 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr11_+_44738699 | 1.10 |
ENSDART00000172301
ENSDART00000170118 |
tk1
|
thymidine kinase 1, soluble |
| chr17_+_18791046 | 1.09 |
ENSDART00000130899
|
si:dkey-288a3.2
|
si:dkey-288a3.2 |
| chr1_-_46260853 | 1.08 |
|
|
|
| chr22_-_6916274 | 1.08 |
|
|
|
| KN150698v1_-_13669 | 1.08 |
|
|
|
| chr17_+_4167546 | 1.07 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr1_+_51118765 | 1.06 |
ENSDART00000156349
|
BX548032.3
|
ENSDARG00000096092 |
| chr21_+_26486084 | 1.06 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr14_-_5100304 | 1.05 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr18_-_35152017 | 1.04 |
|
|
|
| chr22_-_3740362 | 1.04 |
|
|
|
| chr20_+_44684340 | 1.04 |
ENSDART00000149775
|
atad2b
|
ATPase family, AAA domain containing 2B |
| chr10_-_109097 | 1.01 |
ENSDART00000127228
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr9_+_50341338 | 1.00 |
|
|
|
| chr7_-_6273677 | 0.99 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr23_+_38335290 | 0.99 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr25_-_34235633 | 0.97 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr14_+_5921998 | 0.96 |
ENSDART00000098346
|
smu1a
|
smu-1 suppressor of mec-8 and unc-52 homolog a (C. elegans) |
| chr7_-_6328278 | 0.96 |
ENSDART00000130760
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr23_+_22052173 | 0.96 |
ENSDART00000087110
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr25_-_34235582 | 0.95 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr14_-_16469435 | 0.94 |
ENSDART00000113711
|
zgc:123335
|
zgc:123335 |
| chr22_+_1965847 | 0.94 |
|
|
|
| chr14_-_15776937 | 0.94 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr17_-_22553428 | 0.93 |
ENSDART00000079390
|
exo1
|
exonuclease 1 |
| chr8_+_52429114 | 0.93 |
ENSDART00000021604
|
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr1_-_22577855 | 0.92 |
ENSDART00000175685
|
rfc1
|
replication factor C (activator 1) 1 |
| chr6_-_33946853 | 0.92 |
ENSDART00000057290
|
prpf38a
|
pre-mRNA processing factor 38A |
| chr8_+_53734037 | 0.92 |
ENSDART00000179055
|
ENSDARG00000106018
|
ENSDARG00000106018 |
| chr20_-_33584691 | 0.92 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| KN150589v1_-_5247 | 0.91 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr5_-_32655521 | 0.91 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr2_-_29939814 | 0.91 |
ENSDART00000031130
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr2_+_5383347 | 0.91 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr8_+_30103357 | 0.90 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr11_-_25223169 | 0.89 |
|
|
|
| chr25_+_11212787 | 0.89 |
ENSDART00000159583
|
FQ312013.1
|
ENSDARG00000099473 |
| chr17_+_26809555 | 0.89 |
ENSDART00000136971
|
jade1
|
jade family PHD finger 1 |
| chr19_-_20819057 | 0.88 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr21_-_30371822 | 0.88 |
ENSDART00000138384
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr11_-_25223110 | 0.87 |
|
|
|
| chr24_+_17201072 | 0.86 |
ENSDART00000024722
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr2_+_11335733 | 0.86 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr10_+_21477806 | 0.85 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 2.1 | 10.4 | GO:1902103 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 2.0 | 14.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.6 | 6.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 1.3 | 6.4 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.2 | 3.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.0 | 5.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.0 | 3.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.0 | 12.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.8 | 0.8 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.8 | 3.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.7 | 3.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.6 | 2.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.5 | 2.6 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.5 | 1.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.8 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 1.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 2.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.4 | 4.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.4 | 2.0 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.4 | 1.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.3 | 2.1 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.3 | 4.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.3 | 1.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.3 | 1.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.3 | 1.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 0.8 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.2 | 0.7 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.2 | 0.7 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 0.9 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 1.7 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 3.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 0.6 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.2 | 0.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 5.0 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.2 | 0.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.7 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 2.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.9 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 2.7 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 1.9 | GO:0045005 | DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.1 | 2.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 3.8 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.1 | 0.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 2.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.1 | 0.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) negative regulation of histone methylation(GO:0031061) |
| 0.1 | 5.1 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 0.6 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 3.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.3 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 1.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.3 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0043985 | histone arginine methylation(GO:0034969) histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.8 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.4 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 2.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 1.1 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.4 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.8 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 1.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.6 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 6.2 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.6 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 1.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) lipid digestion(GO:0044241) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.9 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 1.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.1 | GO:0008334 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 1.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 5.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.0 | 3.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.6 | 1.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.6 | 3.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.5 | 2.9 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.4 | 3.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 1.7 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.3 | 1.9 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 1.9 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 5.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 0.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.3 | GO:0044304 | main axon(GO:0044304) |
| 0.1 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 1.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 5.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 2.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 1.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 7.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 7.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.3 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.7 | 5.2 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.7 | 5.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.7 | 3.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.7 | 2.8 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.6 | 6.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.6 | 4.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 1.6 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.4 | 1.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 2.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.4 | 1.7 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.4 | 7.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 0.7 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.3 | 1.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 2.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 1.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.3 | 2.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 0.7 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.2 | 2.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 2.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.6 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 4.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 19.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 11.8 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 2.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 8.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 2.7 | GO:0016407 | acetyltransferase activity(GO:0016407) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 2.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.9 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 2.4 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 6.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.3 | 11.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 8.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 9.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 5.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 21.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 1.1 | 11.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 4.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.3 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 3.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 3.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |