Project
DANIO-CODE
Navigation
Downloads
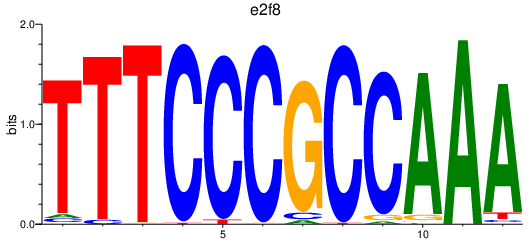
Results for e2f8
Z-value: 0.50
Transcription factors associated with e2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e2f8
|
ENSDARG00000057323 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e2f8 | dr10_dc_chr7_-_16345978_16346013 | 0.65 | 5.9e-03 | Click! |
Activity profile of e2f8 motif
Sorted Z-values of e2f8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of e2f8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_34571082 | 1.25 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr6_+_50452131 | 1.08 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr7_-_55346967 | 0.95 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr10_+_6925373 | 0.83 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr1_-_33717934 | 0.80 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr13_+_46652067 | 0.79 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr23_+_26099718 | 0.72 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr21_-_11562388 | 0.70 |
ENSDART00000139289
|
cast
|
calpastatin |
| chr7_+_24543671 | 0.68 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr4_+_292818 | 0.64 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr22_-_38321005 | 0.64 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr20_+_18803841 | 0.62 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr23_-_33812361 | 0.60 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr13_+_31271568 | 0.58 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr9_+_33409580 | 0.58 |
ENSDART00000100893
|
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr6_+_50452229 | 0.57 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr7_+_41532360 | 0.57 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr16_+_33999554 | 0.56 |
ENSDART00000159969
|
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr22_+_26840870 | 0.55 |
ENSDART00000166775
|
crebbpa
|
CREB binding protein a |
| chr1_-_39141284 | 0.54 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr19_+_43090603 | 0.53 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr1_-_18118467 | 0.53 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr22_+_26840517 | 0.52 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr1_-_37451414 | 0.52 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr11_-_11809024 | 0.52 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr2_-_9854212 | 0.51 |
ENSDART00000112995
|
wu:fi34b01
|
wu:fi34b01 |
| chr16_-_9562744 | 0.49 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr17_+_22291644 | 0.48 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr16_+_25202230 | 0.48 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr16_-_9562608 | 0.47 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr17_+_22291546 | 0.47 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr23_+_29431734 | 0.47 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr24_-_13205000 | 0.45 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr19_+_43090664 | 0.45 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr19_+_14490203 | 0.45 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr3_-_35735085 | 0.44 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr6_+_38898605 | 0.44 |
ENSDART00000029930
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr7_-_38363533 | 0.43 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr23_+_29431388 | 0.43 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr6_+_4000356 | 0.43 |
ENSDART00000111817
|
trim25l
|
tripartite motif containing 25, like |
| chr3_+_25868700 | 0.42 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr8_+_41004169 | 0.41 |
|
|
|
| chr25_+_32114026 | 0.41 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr25_-_34635772 | 0.40 |
ENSDART00000113870
|
ENSDARG00000075379
|
ENSDARG00000075379 |
| chr3_-_41983671 | 0.40 |
ENSDART00000040753
|
nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr5_-_37359524 | 0.40 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr17_+_22291961 | 0.40 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr7_+_26274744 | 0.40 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr23_-_43396787 | 0.40 |
|
|
|
| chr14_-_28260272 | 0.39 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr19_+_40482359 | 0.39 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr16_+_33999263 | 0.38 |
ENSDART00000164447
|
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr16_-_39245193 | 0.37 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr20_-_48957517 | 0.37 |
ENSDART00000158993
|
BX663521.1
|
ENSDARG00000102751 |
| chr18_-_35861412 | 0.36 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr2_-_25464283 | 0.35 |
ENSDART00000099340
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr23_-_17055508 | 0.35 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr21_-_31215463 | 0.34 |
ENSDART00000170507
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr24_-_2830194 | 0.34 |
ENSDART00000164913
|
si:ch211-152c8.5
|
si:ch211-152c8.5 |
| chr12_+_14111747 | 0.34 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr7_+_26274687 | 0.34 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr7_-_41532268 | 0.33 |
ENSDART00000016105
ENSDART00000138800 |
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr7_+_28453707 | 0.33 |
ENSDART00000173947
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr21_-_31215400 | 0.33 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr7_+_23224924 | 0.33 |
ENSDART00000142401
|
zgc:109889
|
zgc:109889 |
| chr6_-_33931989 | 0.33 |
ENSDART00000141483
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr11_+_6209097 | 0.33 |
ENSDART00000156437
|
BX248501.1
|
ENSDARG00000074519 |
| chr25_-_34605630 | 0.32 |
ENSDART00000099859
|
CU302436.2
|
ENSDARG00000068928 |
| chr25_-_34605715 | 0.32 |
ENSDART00000099859
|
CU302436.2
|
ENSDARG00000068928 |
| chr14_+_50232242 | 0.32 |
ENSDART00000110099
|
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr5_-_29782745 | 0.32 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr20_-_21001621 | 0.32 |
ENSDART00000010564
|
xrcc3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr5_-_63773138 | 0.32 |
ENSDART00000172321
|
brd3b
|
bromodomain containing 3b |
| chr6_-_22060441 | 0.31 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr14_-_33010712 | 0.31 |
ENSDART00000010019
|
upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr21_-_11562321 | 0.31 |
ENSDART00000144370
|
cast
|
calpastatin |
| chr7_+_71293928 | 0.31 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr4_-_71846854 | 0.31 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr7_+_5835252 | 0.30 |
ENSDART00000170763
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr3_-_52837780 | 0.30 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr22_-_26254136 | 0.30 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr19_+_43526327 | 0.30 |
|
|
|
| chr3_-_45369628 | 0.30 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr23_-_17055376 | 0.30 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr19_+_43090486 | 0.29 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr7_-_667841 | 0.29 |
ENSDART00000160644
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr23_-_17055330 | 0.29 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr13_+_26573404 | 0.29 |
ENSDART00000142483
|
fancl
|
Fanconi anemia, complementation group L |
| chr7_+_5853590 | 0.28 |
ENSDART00000123660
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr19_-_30771558 | 0.28 |
ENSDART00000103471
|
khdrbs1b
|
KH domain containing, RNA binding, signal transduction associated 1b |
| chr18_-_35861482 | 0.28 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr2_+_9854294 | 0.28 |
|
|
|
| chr4_-_7861030 | 0.28 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr7_+_23224800 | 0.28 |
ENSDART00000115299
ENSDART00000101423 |
zgc:109889
|
zgc:109889 |
| chr7_+_73601671 | 0.28 |
ENSDART00000163075
|
zgc:173552
|
zgc:173552 |
| chr6_-_49323847 | 0.27 |
ENSDART00000155232
|
BX569793.1
|
ENSDARG00000096832 |
| chr20_-_48957424 | 0.27 |
ENSDART00000158993
|
BX663521.1
|
ENSDARG00000102751 |
| chr25_+_34628342 | 0.26 |
ENSDART00000155501
|
zgc:110434
|
zgc:110434 |
| chr5_+_71100066 | 0.26 |
ENSDART00000115182
|
nup214
|
nucleoporin 214 |
| chr23_-_29431197 | 0.26 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr7_-_73631286 | 0.26 |
ENSDART00000129254
|
zgc:173552
|
zgc:173552 |
| chr13_-_35346618 | 0.25 |
ENSDART00000057052
|
slx4ip
|
SLX4 interacting protein |
| chr18_+_45711888 | 0.25 |
ENSDART00000176807
|
depdc7
|
DEP domain containing 7 |
| chr23_-_17055458 | 0.25 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr17_+_14776302 | 0.25 |
ENSDART00000154229
|
RTRAF
|
zgc:56576 |
| chr3_+_14421976 | 0.25 |
ENSDART00000171731
|
znf653
|
zinc finger protein 653 |
| chr13_-_45805308 | 0.25 |
ENSDART00000166442
|
si:ch211-62a1.4
|
si:ch211-62a1.4 |
| chr9_+_46748457 | 0.25 |
|
|
|
| chr7_-_6219627 | 0.25 |
ENSDART00000172898
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| chr21_+_37405864 | 0.24 |
ENSDART00000148956
|
znf346
|
zinc finger protein 346 |
| chr3_-_40060674 | 0.24 |
ENSDART00000175118
|
znf598
|
zinc finger protein 598 |
| chr15_+_27000663 | 0.24 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr21_-_11561978 | 0.24 |
ENSDART00000132699
|
cast
|
calpastatin |
| chr2_+_37441802 | 0.24 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr4_-_1496779 | 0.24 |
ENSDART00000166360
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr13_-_14798134 | 0.24 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr17_+_9918253 | 0.24 |
ENSDART00000169892
|
srp54
|
signal recognition particle 54 |
| chr22_+_25570287 | 0.24 |
|
|
|
| chr6_+_50452529 | 0.23 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr16_-_43107682 | 0.23 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr17_+_11218585 | 0.23 |
ENSDART00000149366
|
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr19_+_42936516 | 0.23 |
ENSDART00000165747
|
BX950855.1
|
ENSDARG00000103815 |
| chr21_-_2334989 | 0.23 |
ENSDART00000171645
|
si:ch73-299h12.9
|
si:ch73-299h12.9 |
| chr11_-_6209253 | 0.23 |
ENSDART00000150199
|
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr5_+_38498787 | 0.23 |
ENSDART00000006079
|
bmp2k
|
BMP2 inducible kinase |
| chr16_+_25202272 | 0.23 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr3_+_52290504 | 0.22 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr5_-_37359835 | 0.22 |
ENSDART00000141059
|
mpzl2b
|
myelin protein zero-like 2b |
| chr11_+_44678488 | 0.21 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr18_+_45711856 | 0.21 |
ENSDART00000077341
|
depdc7
|
DEP domain containing 7 |
| chr2_-_25464332 | 0.21 |
ENSDART00000099340
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr2_+_9957535 | 0.21 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr21_-_30371923 | 0.20 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr3_-_9485481 | 0.20 |
|
|
|
| chr24_-_35673629 | 0.20 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr16_+_53391776 | 0.20 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr17_+_11218461 | 0.20 |
ENSDART00000130975
|
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr24_+_21474677 | 0.20 |
|
|
|
| chr2_+_7014241 | 0.19 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr3_-_26675055 | 0.19 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr16_+_31988496 | 0.19 |
ENSDART00000058496
|
phc1
|
polyhomeotic homolog 1 |
| chr21_-_41804961 | 0.19 |
ENSDART00000046076
|
ENSDARG00000030665
|
ENSDARG00000030665 |
| chr20_+_21578759 | 0.18 |
|
|
|
| chr13_-_26573327 | 0.18 |
ENSDART00000141232
|
BX005477.1
|
ENSDARG00000093327 |
| chr16_+_32230379 | 0.18 |
ENSDART00000084009
|
zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr5_-_29935062 | 0.18 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr11_-_17882817 | 0.17 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr6_+_38898811 | 0.17 |
ENSDART00000131347
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr22_+_26840335 | 0.17 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr16_+_32230439 | 0.17 |
ENSDART00000084009
|
zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr21_+_18369660 | 0.17 |
|
|
|
| chr22_-_3986398 | 0.17 |
ENSDART00000169317
ENSDART00000171262 |
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr5_-_29782972 | 0.16 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr20_+_27193945 | 0.16 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr20_+_53563189 | 0.16 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr1_-_37452112 | 0.16 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr16_+_25201900 | 0.16 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr13_+_7923056 | 0.15 |
ENSDART00000080465
|
hells
|
helicase, lymphoid-specific |
| chr7_-_667670 | 0.15 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr19_-_2651862 | 0.14 |
ENSDART00000113829
|
cdca7b
|
cell division cycle associated 7b |
| chr19_-_18315998 | 0.14 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr11_-_17883113 | 0.14 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr7_-_6219521 | 0.14 |
ENSDART00000161304
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| chr2_+_47885621 | 0.14 |
|
|
|
| chr11_-_11808973 | 0.13 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr9_+_50670635 | 0.13 |
ENSDART00000179475
|
CABZ01083707.1
|
ENSDARG00000107278 |
| chr4_+_73504617 | 0.13 |
ENSDART00000148575
|
nup50
|
nucleoporin 50 |
| chr9_+_50670606 | 0.12 |
ENSDART00000179475
|
CABZ01083707.1
|
ENSDARG00000107278 |
| chr18_+_5260404 | 0.12 |
ENSDART00000150992
|
wdr59
|
WD repeat domain 59 |
| chr15_-_33949567 | 0.12 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr25_-_34640899 | 0.12 |
ENSDART00000157137
|
ENSDARG00000070254
|
ENSDARG00000070254 |
| chr5_-_63773215 | 0.12 |
ENSDART00000168030
|
brd3b
|
bromodomain containing 3b |
| chr13_-_17880164 | 0.12 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr15_+_39400668 | 0.12 |
|
|
|
| chr9_-_2886135 | 0.12 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr24_+_6649249 | 0.12 |
|
|
|
| chr13_-_26573250 | 0.11 |
ENSDART00000139322
|
BX005477.1
|
ENSDARG00000093327 |
| chr21_-_2335024 | 0.11 |
ENSDART00000171645
|
si:ch73-299h12.9
|
si:ch73-299h12.9 |
| chr25_-_26309873 | 0.11 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr13_+_48067433 | 0.11 |
ENSDART00000171080
|
msh6
|
mutS homolog 6 (E. coli) |
| chr14_-_32291093 | 0.11 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase, Class VI, type 11C |
| chr1_-_37452393 | 0.11 |
ENSDART00000047159
|
hmgb2a
|
high mobility group box 2a |
| chr20_+_13245355 | 0.10 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr18_-_5260386 | 0.10 |
ENSDART00000027115
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr5_-_15639015 | 0.10 |
ENSDART00000051644
|
coq5
|
coenzyme Q5, methyltransferase |
| chr21_+_37405904 | 0.10 |
ENSDART00000148956
|
znf346
|
zinc finger protein 346 |
| chr15_-_33949438 | 0.10 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr21_+_37405576 | 0.10 |
ENSDART00000085786
|
znf346
|
zinc finger protein 346 |
| chr15_-_37202518 | 0.10 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr1_+_56656523 | 0.10 |
ENSDART00000152229
|
ENSDARG00000007289
|
ENSDARG00000007289 |
| chr25_-_34604544 | 0.09 |
ENSDART00000154856
|
CU302436.5
|
Histone H3.2 |
| chr5_-_29782945 | 0.09 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr13_-_48068159 | 0.09 |
ENSDART00000165157
|
CU468002.1
|
ENSDARG00000103616 |
| chr15_-_43447148 | 0.09 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr18_+_27593121 | 0.09 |
ENSDART00000134714
|
cd82b
|
CD82 molecule b |
| chr22_-_26254217 | 0.08 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr16_+_7213299 | 0.08 |
ENSDART00000162610
|
bmper
|
BMP binding endothelial regulator |
| chr16_+_53391438 | 0.08 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr3_+_52290252 | 0.08 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr17_-_52233071 | 0.08 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.8 | GO:0009177 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.1 | 1.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 1.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.8 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.3 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.1 | 0.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) temperature homeostasis(GO:0001659) |
| 0.1 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.8 | GO:0008406 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.1 | 0.2 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.6 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic cell cycle, embryonic(GO:0045448) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 1.4 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.6 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 1.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.4 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.3 | GO:0051092 | NIK/NF-kappaB signaling(GO:0038061) positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.2 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.3 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.1 | 0.3 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.1 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.4 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0008425 | 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity(GO:0008425) quinone cofactor methyltransferase activity(GO:0030580) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |