Project
DANIO-CODE
Navigation
Downloads
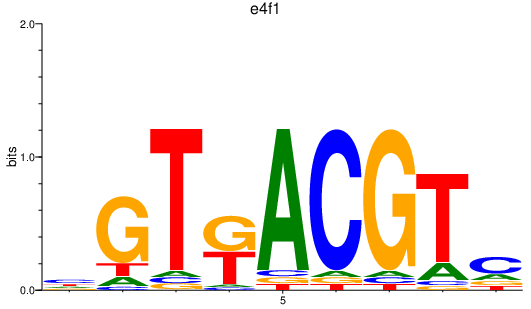
Results for e4f1
Z-value: 0.68
Transcription factors associated with e4f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
e4f1
|
ENSDARG00000038243 | E4F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| e4f1 | dr10_dc_chr3_-_18587352_18587455 | 0.80 | 2.2e-04 | Click! |
Activity profile of e4f1 motif
Sorted Z-values of e4f1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of e4f1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_44539778 | 2.31 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr16_-_41585481 | 1.39 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr5_-_28931727 | 1.35 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr3_+_33608070 | 1.28 |
|
|
|
| chr15_-_23786063 | 1.18 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr25_+_34340569 | 1.15 |
ENSDART00000157638
|
tmem231
|
transmembrane protein 231 |
| chr20_-_44678938 | 1.15 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr2_-_19705537 | 1.05 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr19_+_43090603 | 1.04 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr1_-_54488824 | 0.96 |
ENSDART00000150430
|
pane1
|
proliferation associated nuclear element |
| chr16_-_13723352 | 0.95 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr19_-_44459352 | 0.90 |
ENSDART00000151084
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr9_+_33523707 | 0.90 |
ENSDART00000171649
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr2_+_9762781 | 0.89 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr20_+_16740320 | 0.88 |
ENSDART00000152359
|
tmem30ab
|
transmembrane protein 30Ab |
| chr4_+_1873500 | 0.83 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr22_-_26971170 | 0.83 |
ENSDART00000162742
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr18_+_6417959 | 0.82 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr25_+_30627892 | 0.80 |
|
|
|
| chr25_+_30627755 | 0.78 |
|
|
|
| chr4_+_9010972 | 0.76 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr13_-_7972442 | 0.72 |
ENSDART00000128005
|
atl2
|
atlastin GTPase 2 |
| chr22_-_510040 | 0.70 |
ENSDART00000140101
|
ccnd3
|
cyclin D3 |
| chr19_+_43090664 | 0.70 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr5_+_71360249 | 0.68 |
|
|
|
| chr8_-_19236074 | 0.68 |
ENSDART00000137994
|
zgc:77486
|
zgc:77486 |
| chr23_-_30837726 | 0.68 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr23_+_17583383 | 0.67 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr2_+_9762627 | 0.67 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr2_-_4191540 | 0.66 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr3_-_15337208 | 0.65 |
ENSDART00000011320
|
nfatc2ip
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein |
| chr7_-_24722804 | 0.65 |
|
|
|
| chr17_+_25168837 | 0.65 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr23_-_20199769 | 0.64 |
ENSDART00000147725
|
tktb
|
transketolase b |
| chr23_+_17583666 | 0.64 |
ENSDART00000054738
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr8_-_19235896 | 0.64 |
ENSDART00000036148
|
zgc:77486
|
zgc:77486 |
| chr13_-_7972530 | 0.64 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr3_-_21006698 | 0.63 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr7_+_66410533 | 0.62 |
ENSDART00000027616
ENSDART00000162763 |
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr16_-_13723295 | 0.61 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr10_+_14530451 | 0.61 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr12_+_33793663 | 0.61 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr19_+_43090486 | 0.59 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr12_+_48295188 | 0.59 |
ENSDART00000164427
|
ndr2
|
nodal-related 2 |
| chr17_-_31195299 | 0.58 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| KN150065v1_-_622 | 0.58 |
|
|
|
| chr16_-_41585555 | 0.57 |
ENSDART00000131706
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| KN150030v1_-_22613 | 0.56 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr13_-_36312975 | 0.56 |
ENSDART00000084867
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr24_-_37438115 | 0.56 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr2_-_6380292 | 0.56 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr12_+_33793468 | 0.55 |
ENSDART00000130853
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr16_+_26858575 | 0.55 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr12_-_926596 | 0.53 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| KN150075v1_-_7761 | 0.53 |
ENSDART00000163569
ENSDART00000166885 |
CABZ01094849.1
|
ENSDARG00000103185 |
| chr7_+_66410947 | 0.51 |
ENSDART00000027616
ENSDART00000162763 |
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr17_-_31195376 | 0.51 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr10_-_2944190 | 0.51 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr16_-_13723778 | 0.51 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr4_-_26046562 | 0.51 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr22_-_10511906 | 0.48 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr6_+_94581 | 0.47 |
ENSDART00000125176
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_44539726 | 0.46 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr23_-_30837755 | 0.45 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr10_-_2944295 | 0.45 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr21_-_9689702 | 0.45 |
ENSDART00000158836
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr8_-_19235793 | 0.44 |
ENSDART00000036148
|
zgc:77486
|
zgc:77486 |
| chr20_-_36768780 | 0.44 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr25_-_34340016 | 0.44 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr20_-_36768848 | 0.44 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr15_-_23786239 | 0.43 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr12_+_33219727 | 0.43 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr13_+_17333378 | 0.42 |
ENSDART00000139245
|
CT025897.1
|
ENSDARG00000093577 |
| chr4_-_12931110 | 0.42 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr23_+_17583294 | 0.40 |
ENSDART00000148457
|
gid8b
|
GID complex subunit 8 homolog b (S. cerevisiae) |
| chr2_+_56045684 | 0.38 |
ENSDART00000021011
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr9_+_52308527 | 0.38 |
|
|
|
| chr14_-_38606412 | 0.37 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr16_+_26858600 | 0.36 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr14_+_30073402 | 0.36 |
ENSDART00000136009
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr23_-_30837818 | 0.35 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| KN150213v1_-_1975 | 0.34 |
|
|
|
| chr6_+_59580339 | 0.33 |
|
|
|
| chr16_-_4940519 | 0.33 |
ENSDART00000149421
|
rpa2
|
replication protein A2 |
| chr24_-_37438357 | 0.33 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr12_+_48295296 | 0.32 |
ENSDART00000164427
|
ndr2
|
nodal-related 2 |
| chr13_-_17333382 | 0.30 |
ENSDART00000145499
|
c13h10orf11
|
c13h10orf11 homolog (H. sapiens) |
| chr17_+_8596825 | 0.30 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr4_+_1873543 | 0.30 |
ENSDART00000021139
ENSDART00000144356 |
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr23_-_30837699 | 0.29 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr23_+_2105366 | 0.28 |
|
|
|
| chr22_-_4375781 | 0.28 |
ENSDART00000081969
|
rad23ab
|
RAD23 homolog A, nucleotide excision repair protein b |
| chr13_-_17333287 | 0.28 |
ENSDART00000145499
|
c13h10orf11
|
c13h10orf11 homolog (H. sapiens) |
| chr15_-_34799968 | 0.28 |
ENSDART00000110964
|
bag6
|
BCL2-associated athanogene 6 |
| chr15_+_20303539 | 0.27 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr24_-_37438154 | 0.26 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr1_-_54488896 | 0.26 |
ENSDART00000150430
|
pane1
|
proliferation associated nuclear element |
| chr20_+_36909452 | 0.24 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr15_-_1880173 | 0.24 |
ENSDART00000154175
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_+_7127672 | 0.23 |
ENSDART00000050597
|
xpr1b
|
xenotropic and polytropic retrovirus receptor 1b |
| chr10_+_23130414 | 0.23 |
ENSDART00000022170
|
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr13_-_36313037 | 0.22 |
ENSDART00000084867
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr5_+_35839198 | 0.22 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr14_+_31278635 | 0.21 |
ENSDART00000158875
ENSDART00000026195 |
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr7_-_26246798 | 0.21 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr21_-_1956120 | 0.20 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr19_-_46631565 | 0.20 |
|
|
|
| chr3_+_60988788 | 0.19 |
ENSDART00000110030
|
lmtk2
|
lemur tyrosine kinase 2 |
| chr5_+_36250811 | 0.18 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr15_+_20303673 | 0.17 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr15_-_1880375 | 0.17 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_+_33219444 | 0.17 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr11_-_17648202 | 0.17 |
ENSDART00000080770
|
tmf1
|
TATA element modulatory factor 1 |
| chr23_-_20199837 | 0.17 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr17_-_9839620 | 0.17 |
ENSDART00000148463
|
snx6
|
sorting nexin 6 |
| chr15_+_20303330 | 0.17 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr17_-_32668462 | 0.15 |
ENSDART00000155321
|
CU019637.1
|
ENSDARG00000097530 |
| chr25_+_35540123 | 0.14 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr4_-_12930797 | 0.14 |
ENSDART00000108552
|
lemd3
|
LEM domain containing 3 |
| chr21_-_1956163 | 0.14 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr2_+_56045488 | 0.13 |
ENSDART00000021011
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr6_-_57542175 | 0.13 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr23_-_5825177 | 0.12 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr2_-_19705322 | 0.09 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr10_+_45056951 | 0.08 |
ENSDART00000158553
|
ENSDARG00000101143
|
ENSDARG00000101143 |
| chr1_-_22905093 | 0.07 |
ENSDART00000138852
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr5_-_57045042 | 0.04 |
ENSDART00000141630
|
si:dkey-71l1.1
|
si:dkey-71l1.1 |
| chr10_-_44170848 | 0.04 |
ENSDART00000135240
|
acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr11_+_2641147 | 0.03 |
ENSDART00000132768
|
mapk14b
|
mitogen-activated protein kinase 14b |
| chr18_+_6898118 | 0.01 |
ENSDART00000018735
|
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr19_+_20579471 | 0.00 |
ENSDART00000090942
|
ccdc126
|
coiled-coil domain containing 126 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.3 | 1.1 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 2.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.7 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 0.9 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 3.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 1.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.5 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.1 | 0.4 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.7 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.4 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.6 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 2.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 3.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.6 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.2 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 0.9 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.8 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 0.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.6 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 3.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |