Project
DANIO-CODE
Navigation
Downloads
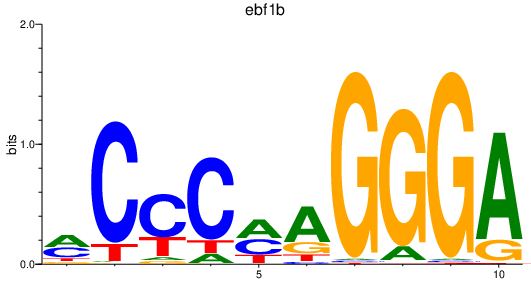
Results for ebf1b
Z-value: 0.84
Transcription factors associated with ebf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf1b
|
ENSDARG00000069196 | EBF transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf1b | dr10_dc_chr21_-_33961137_33961159 | 0.84 | 4.5e-05 | Click! |
Activity profile of ebf1b motif
Sorted Z-values of ebf1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_28814066 | 3.91 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr11_+_6118409 | 2.91 |
ENSDART00000027666
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr7_-_32752138 | 2.54 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr22_+_11726312 | 2.42 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr2_-_30675594 | 2.33 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr13_-_39033893 | 2.12 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr19_+_20183210 | 2.11 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr21_+_26352675 | 2.05 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr9_+_41720118 | 1.88 |
|
|
|
| chr18_+_38773971 | 1.84 |
ENSDART00000010177
|
onecut1
|
one cut homeobox 1 |
| chr10_-_31619761 | 1.83 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr7_-_52283383 | 1.77 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr25_+_11317025 | 1.73 |
|
|
|
| chr25_+_30746445 | 1.66 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr2_+_19546837 | 1.62 |
ENSDART00000163137
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr7_+_25649559 | 1.61 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr25_+_25358457 | 1.56 |
ENSDART00000112330
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr2_+_57136624 | 1.52 |
ENSDART00000159912
|
ENSDARG00000105208
|
ENSDARG00000105208 |
| chr4_-_10600228 | 1.42 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr20_+_30948175 | 1.41 |
|
|
|
| chr2_+_22839514 | 1.33 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 family, member Da |
| chr8_-_15144533 | 1.32 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr11_+_6118322 | 1.32 |
ENSDART00000165031
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_+_1308683 | 1.13 |
ENSDART00000159047
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr12_+_13614102 | 1.10 |
ENSDART00000152689
|
oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr16_-_30606364 | 1.05 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr23_+_23305483 | 1.04 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr7_-_33994315 | 1.02 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr8_+_39525254 | 1.00 |
|
|
|
| chr4_+_13811932 | 0.99 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr14_-_21662462 | 0.98 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr10_+_26156392 | 0.95 |
ENSDART00000079207
|
trim47
|
tripartite motif containing 47 |
| chr23_-_45266310 | 0.94 |
|
|
|
| chr8_-_24276524 | 0.93 |
|
|
|
| chr24_-_28813986 | 0.89 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr8_-_38308535 | 0.88 |
|
|
|
| chr11_-_9479592 | 0.83 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr5_-_57016269 | 0.83 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr10_+_21693418 | 0.82 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr3_-_30027552 | 0.79 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr3_-_38642067 | 0.79 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr11_-_39864306 | 0.79 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr5_-_63912916 | 0.79 |
|
|
|
| chr16_-_21981065 | 0.76 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr24_-_25546068 | 0.76 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr7_-_57727148 | 0.73 |
|
|
|
| chr15_+_42626315 | 0.72 |
|
|
|
| chr3_+_52901602 | 0.71 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr7_+_58384379 | 0.70 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr21_-_18981828 | 0.70 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr13_+_28574593 | 0.69 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr11_-_30792331 | 0.68 |
|
|
|
| chr20_+_30979286 | 0.68 |
|
|
|
| chr13_+_28723589 | 0.67 |
|
|
|
| chr9_-_5618609 | 0.66 |
ENSDART00000127219
|
FAM155A
|
family with sequence similarity 155 member A |
| chr6_+_42478185 | 0.66 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr24_-_25545773 | 0.65 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr22_+_11726429 | 0.61 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr11_-_9479689 | 0.61 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr19_-_27363603 | 0.61 |
|
|
|
| chr2_-_39709905 | 0.60 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr11_-_39864543 | 0.56 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr1_+_44044807 | 0.56 |
ENSDART00000133926
ENSDART00000049701 |
p2rx3b
|
purinergic receptor P2X, ligand-gated ion channel, 3b |
| chr11_-_22143133 | 0.55 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr23_+_18565721 | 0.54 |
|
|
|
| chr25_-_34621069 | 0.47 |
ENSDART00000123892
|
hist1h4l
|
histone 1, H4, like |
| chr24_-_12602773 | 0.47 |
ENSDART00000093583
|
cox19
|
COX19 cytochrome c oxidase assembly factor |
| chr4_+_20596509 | 0.46 |
|
|
|
| chr5_-_63912962 | 0.45 |
|
|
|
| chr13_-_39033849 | 0.44 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr12_+_5283561 | 0.43 |
|
|
|
| chr6_+_48490235 | 0.43 |
ENSDART00000161184
|
si:dkey-238f9.1
|
si:dkey-238f9.1 |
| chr16_+_24047857 | 0.42 |
|
|
|
| chr4_-_10600179 | 0.42 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr24_-_28813917 | 0.42 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr6_+_55091896 | 0.42 |
|
|
|
| chr20_+_48950833 | 0.41 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr10_+_45213089 | 0.41 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| KN150708v1_-_31573 | 0.41 |
ENSDART00000160040
|
slc16a6a
|
solute carrier family 16, member 6a |
| chr5_+_3965645 | 0.40 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr6_-_25393928 | 0.40 |
ENSDART00000160544
|
PKN2 (1 of many)
|
protein kinase N2 |
| chr6_+_15018104 | 0.39 |
ENSDART00000063648
|
nck2b
|
NCK adaptor protein 2b |
| chr25_+_19568627 | 0.38 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr7_+_54347388 | 0.37 |
ENSDART00000158898
|
fgf4
|
fibroblast growth factor 4 |
| chr20_-_23526954 | 0.37 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr16_+_24047649 | 0.36 |
|
|
|
| chr25_+_30473054 | 0.36 |
|
|
|
| chr2_+_43757541 | 0.36 |
|
|
|
| chr1_-_5284684 | 0.35 |
|
|
|
| chr3_-_41944723 | 0.35 |
ENSDART00000083111
|
ttyh3a
|
tweety family member 3a |
| chr7_-_8715924 | 0.34 |
ENSDART00000111002
|
si:ch211-74f19.2
|
si:ch211-74f19.2 |
| chr1_-_32647357 | 0.34 |
ENSDART00000136383
|
cd99
|
CD99 molecule |
| chr3_+_45345181 | 0.33 |
|
|
|
| chr24_-_26594117 | 0.33 |
ENSDART00000124580
|
pld1b
|
phospholipase D1b |
| chr23_+_8261065 | 0.32 |
ENSDART00000044733
|
NPBWR2 (1 of many)
|
neuropeptides B and W receptor 2 |
| chr20_+_31381039 | 0.32 |
|
|
|
| chr24_-_28814284 | 0.31 |
ENSDART00000003503
|
col11a1a
|
collagen, type XI, alpha 1a |
| chr24_-_26594053 | 0.30 |
ENSDART00000124580
|
pld1b
|
phospholipase D1b |
| chr10_-_13438574 | 0.30 |
ENSDART00000030976
|
il11ra
|
interleukin 11 receptor, alpha |
| chr7_-_33690318 | 0.28 |
ENSDART00000100104
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr10_+_19597171 | 0.26 |
ENSDART00000063810
ENSDART00000063806 |
atp6v1b2
|
ATPase, H+ transporting, lysosomal, V1 subunit B2 |
| chr6_+_32428877 | 0.26 |
|
|
|
| chr18_-_15582896 | 0.26 |
ENSDART00000172690
ENSDART00000159915 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr6_-_31268267 | 0.25 |
|
|
|
| chr14_-_6680725 | 0.24 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr4_-_2877346 | 0.24 |
|
|
|
| chr23_-_45266173 | 0.23 |
|
|
|
| chr3_-_19847002 | 0.23 |
|
|
|
| chr19_+_39100820 | 0.21 |
|
|
|
| chr5_+_36332564 | 0.20 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr15_-_1519571 | 0.19 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr6_+_55091976 | 0.19 |
|
|
|
| chr15_-_8786861 | 0.18 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr11_-_39864489 | 0.17 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr8_+_31863748 | 0.16 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr14_-_29498646 | 0.16 |
|
|
|
| chr16_+_24047736 | 0.13 |
|
|
|
| chr24_-_17089437 | 0.13 |
|
|
|
| chr3_-_5318012 | 0.13 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr2_-_22173254 | 0.12 |
ENSDART00000137538
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr15_-_9655500 | 0.12 |
ENSDART00000171214
ENSDART00000171911 |
nars2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr24_-_40962072 | 0.06 |
|
|
|
| chr3_+_62051743 | 0.06 |
ENSDART00000168063
|
zgc:173575
|
zgc:173575 |
| chr7_+_39183037 | 0.05 |
ENSDART00000173520
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr11_+_31295037 | 0.04 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr18_+_18598885 | 0.02 |
|
|
|
| chr12_-_33258663 | 0.00 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.5 | 1.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.5 | 1.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 1.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 2.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 0.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.2 | 1.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 1.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.3 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.6 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 1.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 1.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.7 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.8 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 5.2 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.4 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 1.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.4 | GO:0051781 | positive regulation of cell division(GO:0051781) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 8.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.5 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 4.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.3 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0035381 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 5.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 4.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 2.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.6 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 1.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |