Project
DANIO-CODE
Navigation
Downloads
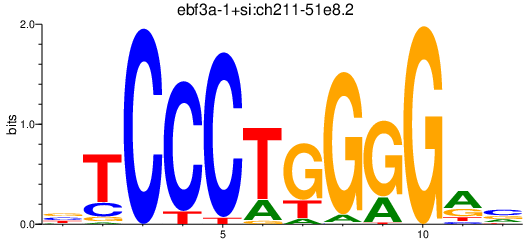
Results for ebf3a-1+si:ch211-51e8.2
Z-value: 0.79
Transcription factors associated with ebf3a-1+si:ch211-51e8.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_ch211-51e8.2
|
ENSDARG00000092635 | si_ch211-51e8.2 |
|
ebf3a
|
ENSDARG00000099849 | EBF transcription factor 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ebf1a | dr10_dc_chr14_+_34683602_34683673 | 0.92 | 5.4e-07 | Click! |
Activity profile of ebf3a-1+si:ch211-51e8.2 motif
Sorted Z-values of ebf3a-1+si:ch211-51e8.2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf3a-1+si:ch211-51e8.2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_32752138 | 3.20 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr22_+_11726312 | 3.00 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr16_-_9978112 | 2.79 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr8_-_985673 | 2.66 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr17_-_45383925 | 2.41 |
|
|
|
| chr25_+_30746445 | 2.40 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr19_+_20183210 | 2.04 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr22_+_11745592 | 1.97 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr25_+_25358457 | 1.96 |
ENSDART00000112330
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr21_-_15833030 | 1.85 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr4_-_76594677 | 1.75 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr24_-_23797468 | 1.70 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr21_+_26352675 | 1.67 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr11_-_22966274 | 1.66 |
ENSDART00000003646
|
optc
|
opticin |
| chr13_+_11913290 | 1.57 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_-_36853345 | 1.57 |
ENSDART00000172074
|
NINJ1
|
ninjurin 1 |
| chr2_+_57136624 | 1.54 |
ENSDART00000159912
|
ENSDARG00000105208
|
ENSDARG00000105208 |
| chr14_-_21662462 | 1.50 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr10_-_31619761 | 1.49 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr7_+_25649559 | 1.44 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr6_+_54703206 | 1.38 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr4_+_8637319 | 1.30 |
|
|
|
| chr12_-_26314881 | 1.29 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr3_+_48362748 | 1.26 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr7_-_52283383 | 1.25 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr2_-_23516930 | 1.24 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr5_-_28090077 | 1.19 |
|
|
|
| chr22_+_11745657 | 1.17 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr6_+_2849460 | 1.16 |
|
|
|
| chr2_-_20490037 | 1.14 |
ENSDART00000160388
|
FQ377605.1
|
ENSDARG00000101927 |
| chr15_-_4154407 | 1.12 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr8_-_15144533 | 1.10 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr8_+_39525254 | 1.05 |
|
|
|
| chr2_-_23517033 | 1.02 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr18_+_37291448 | 0.99 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr20_+_30948175 | 0.97 |
|
|
|
| chr15_-_31635737 | 0.96 |
ENSDART00000156047
|
hmgb1b
|
high mobility group box 1b |
| chr10_+_35478939 | 0.94 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr4_-_16356071 | 0.91 |
ENSDART00000079521
|
kera
|
keratocan |
| chr8_-_27496134 | 0.90 |
ENSDART00000134879
|
si:ch211-254n4.3
|
si:ch211-254n4.3 |
| chr21_+_10663517 | 0.89 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr2_+_22839514 | 0.89 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 family, member Da |
| chr15_-_24934442 | 0.89 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr13_+_28723589 | 0.85 |
|
|
|
| chr20_+_38298893 | 0.85 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr4_-_76594399 | 0.84 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr13_+_28574593 | 0.82 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr19_+_11061965 | 0.82 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr16_-_31835463 | 0.81 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr1_-_11707288 | 0.81 |
ENSDART00000134708
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr22_+_11726429 | 0.79 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr3_-_30027552 | 0.79 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr20_+_30979286 | 0.78 |
|
|
|
| chr13_-_40190349 | 0.78 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr12_+_30245620 | 0.78 |
ENSDART00000153116
ENSDART00000152900 |
ENSDARG00000020224
|
ENSDARG00000020224 |
| chr6_+_42478185 | 0.76 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr5_-_5338903 | 0.74 |
ENSDART00000112856
|
ENSDARG00000078362
|
ENSDARG00000078362 |
| chr20_-_25839801 | 0.73 |
|
|
|
| chr7_+_39139248 | 0.69 |
ENSDART00000165594
|
CT030188.1
|
ENSDARG00000099385 |
| chr20_-_23539928 | 0.69 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr8_+_24276667 | 0.68 |
|
|
|
| chr21_-_9863186 | 0.67 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr1_-_46292847 | 0.66 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr20_+_38298755 | 0.65 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr8_+_24276581 | 0.64 |
|
|
|
| chr19_-_46396376 | 0.64 |
|
|
|
| chr25_-_29245587 | 0.61 |
|
|
|
| chr15_+_24077452 | 0.60 |
|
|
|
| chr24_-_26594117 | 0.59 |
ENSDART00000124580
|
pld1b
|
phospholipase D1b |
| chr4_+_27141061 | 0.59 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr7_-_18302231 | 0.59 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr12_-_425381 | 0.58 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr10_-_27087472 | 0.55 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr24_-_26594053 | 0.54 |
ENSDART00000124580
|
pld1b
|
phospholipase D1b |
| chr11_-_38278666 | 0.53 |
|
|
|
| chr12_+_34662699 | 0.53 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr16_-_7310925 | 0.50 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr3_+_52901602 | 0.49 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr20_+_27121126 | 0.49 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr23_-_36319185 | 0.49 |
ENSDART00000139328
|
znf740b
|
zinc finger protein 740b |
| chr18_+_6692267 | 0.49 |
|
|
|
| chr21_-_9863071 | 0.48 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr7_-_41209891 | 0.48 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr20_+_230164 | 0.46 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr10_-_17383380 | 0.45 |
|
|
|
| chr17_-_120568 | 0.45 |
|
|
|
| chr14_+_41519751 | 0.42 |
ENSDART00000173264
|
slc7a11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr16_-_25691856 | 0.42 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr3_+_12132746 | 0.41 |
|
|
|
| chr6_-_8029184 | 0.41 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr2_-_23516991 | 0.40 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr17_+_24421150 | 0.39 |
ENSDART00000168926
|
mdh1ab
|
malate dehydrogenase 1Ab, NAD (soluble) |
| chr13_-_47510290 | 0.39 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr17_-_120755 | 0.35 |
|
|
|
| chr11_+_2211050 | 0.34 |
ENSDART00000123944
|
hoxc13b
|
homeobox C13b |
| chr10_-_17383466 | 0.34 |
|
|
|
| chr17_-_20538582 | 0.33 |
|
|
|
| chr4_+_15820977 | 0.32 |
ENSDART00000101613
|
lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr5_-_63912962 | 0.31 |
|
|
|
| chr1_-_52869817 | 0.31 |
ENSDART00000162025
ENSDART00000100788 |
commd1
|
copper metabolism (Murr1) domain containing 1 |
| chr16_+_40396454 | 0.30 |
ENSDART00000121711
|
BX072576.2
|
ENSDARG00000090181 |
| chr17_+_33862300 | 0.28 |
|
|
|
| chr24_-_23797593 | 0.28 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr4_-_72835088 | 0.24 |
ENSDART00000174361
|
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr7_+_60746590 | 0.23 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr25_-_17406938 | 0.23 |
ENSDART00000061726
|
trim35-40
|
tripartite motif containing 35-40 |
| chr8_-_17692185 | 0.23 |
|
|
|
| chr15_+_2495106 | 0.23 |
|
|
|
| chr2_-_23516868 | 0.23 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr20_-_48255596 | 0.22 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr14_+_10795333 | 0.22 |
|
|
|
| chr25_-_23977017 | 0.21 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr22_-_27274035 | 0.20 |
ENSDART00000153589
|
si:dkey-16m19.1
|
si:dkey-16m19.1 |
| chr12_+_45515755 | 0.18 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr5_-_32861219 | 0.18 |
ENSDART00000162290
|
ENSDARG00000060631
|
ENSDARG00000060631 |
| chr16_-_45888127 | 0.18 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr7_+_34216359 | 0.18 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr12_+_5283561 | 0.17 |
|
|
|
| chr3_-_19847002 | 0.17 |
|
|
|
| chr12_+_6161471 | 0.16 |
ENSDART00000163828
|
BX248126.1
|
ENSDARG00000096552 |
| chr3_-_55397643 | 0.15 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr22_-_2828572 | 0.12 |
|
|
|
| chr25_+_29245654 | 0.12 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr16_-_42857240 | 0.10 |
|
|
|
| chr7_+_39183037 | 0.10 |
ENSDART00000173520
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr24_+_25649220 | 0.09 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_+_44265618 | 0.08 |
ENSDART00000171690
ENSDART00000161949 |
CT027744.2
|
ENSDARG00000102431 |
| chr1_-_58183299 | 0.07 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr5_-_32860888 | 0.06 |
ENSDART00000085636
|
ENSDARG00000060631
|
ENSDARG00000060631 |
| chr3_-_55397592 | 0.06 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr15_+_28335293 | 0.06 |
ENSDART00000152536
|
myo1cb
|
myosin Ic, paralog b |
| chr1_-_53465275 | 0.05 |
|
|
|
| chr12_-_31369772 | 0.04 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr4_-_16356247 | 0.02 |
ENSDART00000079521
|
kera
|
keratocan |
| chr23_-_16765933 | 0.01 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0086066 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.6 | 1.7 | GO:0019731 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.5 | 2.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.5 | 1.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.4 | 1.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.4 | 2.5 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 0.9 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.3 | 0.9 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 1.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 1.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 1.1 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 0.6 | GO:0051701 | interaction with host(GO:0051701) |
| 0.2 | 1.1 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.2 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 1.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.5 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.5 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.1 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.3 | GO:2000009 | negative regulation of sodium ion transmembrane transport(GO:1902306) regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.2 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.1 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 1.6 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.5 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 2.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 6.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.9 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 1.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 1.5 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 2.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 7.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 1.4 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |