Project
DANIO-CODE
Navigation
Downloads
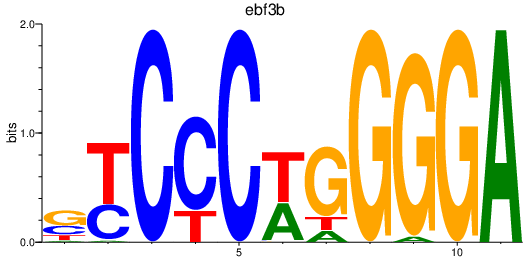
Results for ebf3b
Z-value: 1.57
Transcription factors associated with ebf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ebf3b
|
ENSDARG00000091287 | EBF transcription factor 3b |
Activity profile of ebf3b motif
Sorted Z-values of ebf3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ebf3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_32752138 | 7.42 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr4_+_8637319 | 5.81 |
|
|
|
| chr22_+_11726312 | 5.69 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr25_+_30746445 | 5.30 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr16_-_9978112 | 4.92 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr6_+_54703206 | 4.65 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr19_-_8961733 | 4.64 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr21_-_37509454 | 4.30 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr17_-_45383925 | 4.08 |
|
|
|
| chr22_+_11745592 | 3.97 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_+_20183210 | 3.91 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr11_-_38843219 | 3.67 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr24_-_23797468 | 3.64 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr13_+_371940 | 3.63 |
ENSDART00000013007
|
degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr9_-_7695437 | 3.38 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr8_-_985673 | 3.37 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr6_+_2849460 | 3.34 |
|
|
|
| chr7_-_52283383 | 3.13 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr17_+_17935213 | 2.90 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr12_+_31558667 | 2.70 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr2_-_39710140 | 2.61 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr6_-_49674729 | 2.45 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr7_-_18302231 | 2.37 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr7_-_72127164 | 2.34 |
ENSDART00000176069
|
CABZ01057364.1
|
ENSDARG00000106736 |
| chr10_-_5016621 | 2.34 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr18_+_9413588 | 2.33 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr10_+_33935945 | 2.26 |
|
|
|
| chr8_-_15144533 | 2.19 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_-_62116975 | 2.18 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr4_-_9007828 | 2.12 |
ENSDART00000153776
|
scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr17_+_38619041 | 2.11 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr7_-_33678058 | 2.08 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr4_-_28346021 | 2.04 |
ENSDART00000178149
|
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr21_+_4964891 | 1.99 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr21_+_21337628 | 1.94 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr9_-_31467299 | 1.86 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr13_-_31491759 | 1.80 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr24_+_4946542 | 1.79 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr10_-_31619761 | 1.75 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr23_-_36319185 | 1.75 |
ENSDART00000139328
|
znf740b
|
zinc finger protein 740b |
| chr15_-_24934442 | 1.74 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr22_-_20361311 | 1.71 |
|
|
|
| chr20_+_38298893 | 1.64 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr8_+_39525254 | 1.63 |
|
|
|
| chr8_+_24276667 | 1.56 |
|
|
|
| chr11_-_30792331 | 1.56 |
|
|
|
| chr19_+_11061965 | 1.53 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr20_+_6600184 | 1.53 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr8_-_38308535 | 1.52 |
|
|
|
| chr24_+_4946079 | 1.51 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr22_-_23238714 | 1.51 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr22_+_11726429 | 1.50 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr22_+_11745657 | 1.49 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr11_-_38278666 | 1.48 |
|
|
|
| chr13_-_40190349 | 1.47 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr18_+_6692267 | 1.47 |
|
|
|
| chr8_+_24276581 | 1.42 |
|
|
|
| chr16_-_38604326 | 1.39 |
ENSDART00000109124
|
rspo2
|
R-spondin 2 |
| chr11_-_22143133 | 1.37 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr12_+_34662699 | 1.36 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr6_+_45930323 | 1.32 |
ENSDART00000154896
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr16_-_32059848 | 1.31 |
ENSDART00000138701
|
gstk1
|
glutathione S-transferase kappa 1 |
| chr4_-_38406131 | 1.31 |
|
|
|
| chr20_+_6600246 | 1.28 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr11_-_27253835 | 1.25 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr20_+_38298755 | 1.22 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr3_+_43010408 | 1.21 |
ENSDART00000169061
|
CU138533.1
|
ENSDARG00000099842 |
| chr2_-_39709905 | 1.11 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr13_+_28723589 | 1.03 |
|
|
|
| chr5_-_18548376 | 1.01 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr11_+_7570027 | 0.96 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr7_+_39139248 | 0.96 |
ENSDART00000165594
|
CT030188.1
|
ENSDARG00000099385 |
| chr10_+_8697704 | 0.95 |
ENSDART00000123131
|
pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr22_-_23518719 | 0.94 |
ENSDART00000161253
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr7_+_54347388 | 0.93 |
ENSDART00000158898
|
fgf4
|
fibroblast growth factor 4 |
| chr16_+_40396454 | 0.89 |
ENSDART00000121711
|
BX072576.2
|
ENSDARG00000090181 |
| chr4_-_48343469 | 0.83 |
ENSDART00000121664
|
znf1128
|
zinc finger protein 1128 |
| chr13_+_46487890 | 0.82 |
|
|
|
| chr21_-_37509396 | 0.82 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr4_+_67334424 | 0.77 |
ENSDART00000136092
|
si:ch211-209j12.3
|
si:ch211-209j12.3 |
| chr9_-_37114484 | 0.76 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr3_-_28534446 | 0.76 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr18_+_17552191 | 0.75 |
|
|
|
| chr25_-_21618526 | 0.75 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
dedicator of cytokinesis 4 |
| chr11_+_7570097 | 0.72 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr17_-_43409302 | 0.72 |
ENSDART00000156206
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr22_-_2828572 | 0.70 |
|
|
|
| chr1_-_52869817 | 0.68 |
ENSDART00000162025
ENSDART00000100788 |
commd1
|
copper metabolism (Murr1) domain containing 1 |
| chr24_-_23797593 | 0.64 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr17_-_43409222 | 0.56 |
ENSDART00000156033
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr3_-_55397643 | 0.55 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr3_-_19847002 | 0.54 |
|
|
|
| chr3_-_55397592 | 0.54 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
| chr4_-_9007909 | 0.51 |
ENSDART00000164632
|
scube1
|
signal peptide, CUB domain, EGF-like 1 |
| chr18_+_9413668 | 0.48 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr15_+_2495106 | 0.46 |
|
|
|
| chr9_-_31467205 | 0.44 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr7_+_38540519 | 0.43 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr8_+_31812204 | 0.40 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr4_-_29003925 | 0.36 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr25_+_34707504 | 0.31 |
|
|
|
| chr7_+_60746590 | 0.29 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr17_+_30506560 | 0.27 |
|
|
|
| chr25_-_23977017 | 0.25 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr12_+_6161471 | 0.24 |
ENSDART00000163828
|
BX248126.1
|
ENSDARG00000096552 |
| chr23_+_35573740 | 0.24 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr16_-_42857240 | 0.21 |
|
|
|
| chr21_-_36495488 | 0.20 |
|
|
|
| chr4_+_29775735 | 0.20 |
ENSDART00000166510
|
BX530097.1
|
ENSDARG00000104762 |
| chr6_+_7093685 | 0.18 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr23_-_35904414 | 0.15 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr7_+_39139117 | 0.13 |
ENSDART00000165594
|
CT030188.1
|
ENSDARG00000099385 |
| chr10_+_33936073 | 0.10 |
|
|
|
| chr15_+_2495159 | 0.08 |
|
|
|
| chr8_-_17692185 | 0.07 |
|
|
|
| chr1_+_15855776 | 0.01 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 1.1 | 4.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.8 | 4.7 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.8 | 1.5 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.6 | 2.8 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.5 | 3.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.5 | 2.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 3.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 1.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 4.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 2.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 1.8 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 2.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 | 2.2 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 0.7 | GO:2000008 | negative regulation of sodium ion transmembrane transport(GO:1902306) regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 3.7 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 0.9 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.1 | 2.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.4 | GO:0033336 | caudal fin development(GO:0033336) |
| 0.1 | 1.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 5.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 0.3 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.1 | 2.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.1 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 1.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.6 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 1.3 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.9 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 3.4 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.8 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 2.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 3.1 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 4.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 12.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.0 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 4.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 3.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 4.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.8 | GO:0030054 | cell junction(GO:0030054) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.4 | 2.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 5.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 2.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 3.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 5.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 4.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 10.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 3.6 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 13.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 24.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 6.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |