Project
DANIO-CODE
Navigation
Downloads
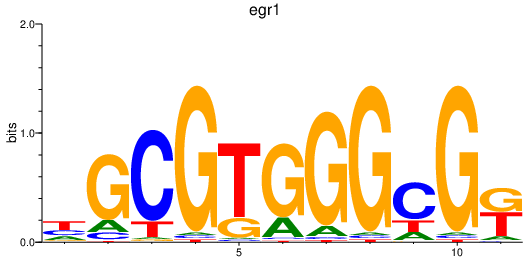
Results for egr1
Z-value: 0.55
Transcription factors associated with egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr1
|
ENSDARG00000037421 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr1 | dr10_dc_chr14_-_21039158_21039162 | -0.25 | 3.6e-01 | Click! |
Activity profile of egr1 motif
Sorted Z-values of egr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of egr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_48188313 | 0.87 |
|
|
|
| chr25_-_35760878 | 0.78 |
ENSDART00000121786
|
hist1h4l
|
histone 1, H4, like |
| chr25_+_35765309 | 0.76 |
ENSDART00000128547
|
hist1h4l
|
histone 1, H4, like |
| chr19_-_6274458 | 0.73 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr14_+_14850200 | 0.70 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr25_-_34634503 | 0.68 |
ENSDART00000157370
|
CR762436.3
|
Histone H3.2 |
| chr23_-_22146031 | 0.66 |
ENSDART00000054362
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr7_-_6273677 | 0.64 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr10_-_34058331 | 0.60 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr25_+_1511300 | 0.59 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr19_-_6274040 | 0.53 |
ENSDART00000140347
ENSDART00000092656 |
erf
|
Ets2 repressor factor |
| KN150256v1_-_9560 | 0.51 |
|
|
|
| chr23_-_22145900 | 0.46 |
ENSDART00000132343
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr19_-_6274169 | 0.46 |
ENSDART00000140347
ENSDART00000092656 |
erf
|
Ets2 repressor factor |
| chr7_-_20358730 | 0.45 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr7_+_5846555 | 0.42 |
ENSDART00000083397
|
hist1h4l
|
histone 1, H4, like |
| chr20_-_54591757 | 0.41 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr6_+_48188448 | 0.40 |
|
|
|
| chr15_-_17163526 | 0.39 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr7_-_52223087 | 0.38 |
ENSDART00000129769
|
cgnl1
|
cingulin-like 1 |
| chr23_-_29630277 | 0.37 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr22_-_62082 | 0.37 |
ENSDART00000166010
ENSDART00000125700 |
mrpl20
|
mitochondrial ribosomal protein L20 |
| chr21_+_19332231 | 0.37 |
ENSDART00000168728
|
btc
|
betacellulin, epidermal growth factor family member |
| chr19_+_1298795 | 0.36 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr5_+_65491173 | 0.34 |
ENSDART00000050847
ENSDART00000172117 |
GLDC
|
glycine decarboxylase |
| chr5_+_37120604 | 0.34 |
ENSDART00000076083
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr16_-_26258657 | 0.34 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr16_-_12621624 | 0.33 |
ENSDART00000155829
|
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr1_-_17000212 | 0.33 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr21_+_19332111 | 0.33 |
ENSDART00000158471
|
btc
|
betacellulin, epidermal growth factor family member |
| chr19_+_7235553 | 0.32 |
ENSDART00000160894
|
brd2a
|
bromodomain containing 2a |
| chr16_-_6881218 | 0.32 |
ENSDART00000149070
|
mbpb
|
myelin basic protein b |
| chr21_-_804453 | 0.31 |
|
|
|
| chr12_-_29118423 | 0.31 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr9_-_41608298 | 0.31 |
|
|
|
| chr6_+_48188520 | 0.31 |
|
|
|
| chr7_-_24373315 | 0.30 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr1_-_48790379 | 0.30 |
ENSDART00000150842
|
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr16_-_5205600 | 0.30 |
ENSDART00000148955
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr2_+_35898064 | 0.28 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr24_+_24777962 | 0.27 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr1_-_51870424 | 0.27 |
ENSDART00000135636
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr8_-_53584884 | 0.27 |
|
|
|
| chr24_+_3296536 | 0.27 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr19_-_6274237 | 0.26 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr12_-_37274632 | 0.25 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr3_+_14151127 | 0.25 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr14_+_891448 | 0.25 |
ENSDART00000021346
|
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr10_+_45184723 | 0.24 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr8_-_49227143 | 0.24 |
|
|
|
| chr3_-_45358502 | 0.24 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr6_+_42340776 | 0.24 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr5_+_65491288 | 0.24 |
ENSDART00000050847
ENSDART00000172117 |
GLDC
|
glycine decarboxylase |
| chr20_+_13998066 | 0.24 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr23_+_2614658 | 0.23 |
|
|
|
| chr7_+_28453707 | 0.23 |
ENSDART00000173947
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr14_-_15393962 | 0.23 |
ENSDART00000161123
ENSDART00000167146 |
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr19_-_25843036 | 0.22 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr8_+_8889274 | 0.22 |
ENSDART00000142075
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr19_-_7235125 | 0.21 |
|
|
|
| chr2_+_38959335 | 0.21 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr3_-_37429712 | 0.21 |
ENSDART00000161401
|
arf2a
|
ADP-ribosylation factor 2a |
| chr19_-_25842574 | 0.21 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr7_-_24724452 | 0.21 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr12_-_2638179 | 0.21 |
|
|
|
| chr9_-_55878791 | 0.21 |
|
|
|
| chr24_+_24778026 | 0.20 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr3_-_5318012 | 0.20 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr6_-_18019342 | 0.20 |
ENSDART00000154941
|
ten1
|
TEN1 CST complex subunit |
| chr2_+_38288916 | 0.19 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr8_+_20406797 | 0.19 |
ENSDART00000016422
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr18_+_15137751 | 0.19 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr2_+_24046083 | 0.19 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr2_+_50327729 | 0.19 |
ENSDART00000097939
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr19_-_2147899 | 0.19 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr8_-_25889113 | 0.18 |
|
|
|
| chr11_+_25058986 | 0.18 |
ENSDART00000122249
|
top1l
|
topoisomerase (DNA) I, like |
| chr1_-_17000175 | 0.18 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr6_-_12972427 | 0.18 |
ENSDART00000147851
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr18_+_36620417 | 0.18 |
ENSDART00000088421
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr20_+_32620786 | 0.18 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr14_+_17092009 | 0.18 |
ENSDART00000129838
|
rnf212
|
ring finger protein 212 |
| chr16_-_5205633 | 0.18 |
ENSDART00000082071
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr17_+_15975348 | 0.17 |
ENSDART00000160742
|
clmn
|
calmin |
| chr22_+_37688695 | 0.17 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr8_-_18582786 | 0.17 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr7_-_10318801 | 0.16 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr15_+_14925920 | 0.16 |
|
|
|
| chr22_+_24112851 | 0.16 |
|
|
|
| chr16_-_14075348 | 0.16 |
ENSDART00000138304
|
si:dkey-85k15.4
|
si:dkey-85k15.4 |
| chr11_-_2524380 | 0.15 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr7_+_22710714 | 0.15 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr7_+_41060622 | 0.15 |
ENSDART00000160143
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr22_+_37689055 | 0.15 |
|
|
|
| chr17_-_1524813 | 0.15 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr18_+_5637539 | 0.15 |
|
|
|
| chr5_+_37120548 | 0.15 |
ENSDART00000076083
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr20_+_27188382 | 0.14 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr21_-_776741 | 0.14 |
|
|
|
| chr20_+_38555185 | 0.14 |
ENSDART00000020153
|
adck3
|
aarF domain containing kinase 3 |
| chr11_+_38449192 | 0.14 |
ENSDART00000041800
|
epha8
|
eph receptor A8 |
| KN150034v1_+_1223 | 0.14 |
|
|
|
| chr2_-_44491143 | 0.14 |
ENSDART00000084174
|
lig1
|
ligase I, DNA, ATP-dependent |
| chr8_-_49227286 | 0.13 |
|
|
|
| chr18_-_39797167 | 0.13 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr17_+_1525358 | 0.13 |
|
|
|
| chr24_+_37800773 | 0.13 |
ENSDART00000137017
|
h3f3d
|
H3 histone, family 3D |
| chr23_+_25929969 | 0.13 |
ENSDART00000145426
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr9_+_33310981 | 0.13 |
|
|
|
| chr8_+_2583789 | 0.13 |
ENSDART00000143242
|
naif1
|
nuclear apoptosis inducing factor 1 |
| chr16_+_19223683 | 0.13 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr14_+_29428950 | 0.13 |
ENSDART00000105898
|
ENSDARG00000071567
|
ENSDARG00000071567 |
| chr15_-_21003820 | 0.12 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr9_+_52256434 | 0.12 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr18_+_45866556 | 0.12 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr22_+_37688988 | 0.12 |
|
|
|
| chr7_+_41060426 | 0.12 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| KN149779v1_-_14268 | 0.12 |
ENSDART00000162865
|
ift27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr24_+_37518715 | 0.12 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr14_+_300285 | 0.12 |
ENSDART00000159949
|
wdr1
|
WD repeat domain 1 |
| chr5_+_6308337 | 0.12 |
ENSDART00000048201
|
SMAD4 (1 of many)
|
SMAD family member 4 |
| chr17_+_26736073 | 0.11 |
ENSDART00000163886
|
larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr23_-_33692244 | 0.11 |
|
|
|
| chr2_+_17042235 | 0.11 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr7_-_20358696 | 0.11 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr7_-_24373258 | 0.11 |
ENSDART00000172282
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr20_-_45757288 | 0.11 |
ENSDART00000124582
ENSDART00000100290 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr8_-_53508979 | 0.10 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr7_-_20358841 | 0.10 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr7_+_45747395 | 0.10 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr23_-_31719203 | 0.10 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_+_24045890 | 0.10 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr3_-_5318147 | 0.10 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr25_+_29245654 | 0.10 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr16_+_14075878 | 0.09 |
ENSDART00000059926
|
zgc:162509
|
zgc:162509 |
| chr6_+_48188609 | 0.09 |
|
|
|
| chr23_+_25930072 | 0.09 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr5_+_37120337 | 0.09 |
ENSDART00000076083
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr5_+_36431907 | 0.09 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr10_-_32907793 | 0.09 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr11_-_37613237 | 0.09 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr20_+_32620873 | 0.09 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr15_-_4491042 | 0.09 |
ENSDART00000157273
|
tfdp2
|
transcription factor Dp-2 |
| chr11_+_36984616 | 0.09 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr3_+_51430252 | 0.09 |
ENSDART00000159493
|
baiap2a
|
BAI1-associated protein 2a |
| chr23_-_32173435 | 0.09 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr1_+_39357275 | 0.09 |
|
|
|
| chr2_-_24947660 | 0.09 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| KN150034v1_+_1158 | 0.08 |
|
|
|
| chr15_+_17471018 | 0.08 |
ENSDART00000145216
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
| chr12_-_20181519 | 0.08 |
ENSDART00000040909
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr7_-_24373522 | 0.08 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr22_+_37688585 | 0.08 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr16_-_14075931 | 0.08 |
|
|
|
| chr23_+_2847589 | 0.08 |
ENSDART00000156954
|
plcg1
|
phospholipase C, gamma 1 |
| chr15_-_17163487 | 0.08 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr7_-_20358897 | 0.08 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr1_-_9789386 | 0.08 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr11_+_269859 | 0.08 |
ENSDART00000109091
|
mettl1
|
methyltransferase like 1 |
| chr13_+_290930 | 0.08 |
ENSDART00000017854
|
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr18_+_5456221 | 0.08 |
|
|
|
| chr5_+_36010806 | 0.07 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr7_+_9080691 | 0.07 |
ENSDART00000104536
|
chsy1
|
chondroitin sulfate synthase 1 |
| chr11_+_25058714 | 0.07 |
ENSDART00000065949
|
top1l
|
topoisomerase (DNA) I, like |
| chr12_-_4212149 | 0.07 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr9_-_33116897 | 0.07 |
|
|
|
| chr18_-_19114558 | 0.07 |
ENSDART00000177621
|
dennd4a
|
DENN/MADD domain containing 4A |
| chr24_-_37800277 | 0.07 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr5_+_35913494 | 0.07 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr19_+_341775 | 0.07 |
ENSDART00000151013
|
ensaa
|
endosulfine alpha a |
| chr15_+_14925529 | 0.07 |
ENSDART00000166250
|
CR936977.2
|
ENSDARG00000103638 |
| chr15_-_23484754 | 0.07 |
ENSDART00000148840
|
kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr11_+_44916426 | 0.06 |
|
|
|
| chr2_-_54909304 | 0.06 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short-chain family member 2 like |
| chr2_-_55565099 | 0.06 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr15_+_42441012 | 0.06 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr5_-_1734159 | 0.05 |
ENSDART00000145781
|
ENSDARG00000094300
|
ENSDARG00000094300 |
| chr2_+_21698627 | 0.05 |
ENSDART00000133228
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr12_-_954375 | 0.05 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr9_-_54459836 | 0.05 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr24_-_42193714 | 0.05 |
|
|
|
| chr9_+_52256395 | 0.05 |
ENSDART00000125850
|
cd302
|
CD302 molecule |
| chr20_+_32620937 | 0.05 |
ENSDART00000147319
|
scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr6_+_12972934 | 0.04 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr18_+_15137711 | 0.04 |
ENSDART00000128609
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr25_-_29245587 | 0.04 |
|
|
|
| chr3_-_23620163 | 0.04 |
|
|
|
| chr9_-_30553051 | 0.04 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr1_-_44239584 | 0.04 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr19_-_2148053 | 0.04 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr2_-_1752776 | 0.04 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr16_-_13704905 | 0.04 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr21_+_8106096 | 0.03 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr23_+_24557956 | 0.03 |
|
|
|
| chr18_-_5689483 | 0.03 |
ENSDART00000129459
|
spata5l1
|
spermatogenesis associated 5-like 1 |
| chr6_-_25081182 | 0.03 |
ENSDART00000161585
|
lrrc8db
|
leucine rich repeat containing 8 family, member Db |
| chr20_+_46795264 | 0.03 |
ENSDART00000167398
|
wdr26b
|
WD repeat domain 26b |
| chr7_-_45747112 | 0.03 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr14_+_38505984 | 0.03 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr15_+_17470879 | 0.03 |
ENSDART00000081059
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.3 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.7 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.2 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.8 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.5 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.0 | 0.1 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.2 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.2 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 2.0 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0035999 | folic acid-containing compound biosynthetic process(GO:0009396) tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.1 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.1 | 0.5 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 2.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 0.6 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |