Project
DANIO-CODE
Navigation
Downloads
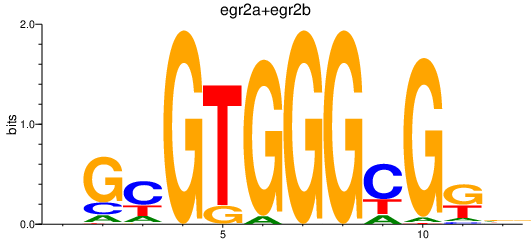
Results for egr2a+egr2b
Z-value: 0.84
Transcription factors associated with egr2a+egr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr2b
|
ENSDARG00000042826 | early growth response 2b |
|
egr2a
|
ENSDARG00000044098 | early growth response 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr2b | dr10_dc_chr12_-_8448482_8448512 | 0.70 | 2.6e-03 | Click! |
Activity profile of egr2a+egr2b motif
Sorted Z-values of egr2a+egr2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of egr2a+egr2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_24063849 | 2.15 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr23_+_20496283 | 2.08 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr9_-_41608298 | 1.90 |
|
|
|
| chr8_-_53508979 | 1.77 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr12_+_11042472 | 1.57 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr25_+_24152717 | 1.54 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr7_-_32752138 | 1.49 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| KN150495v1_-_20497 | 1.44 |
|
|
|
| chr25_+_33527870 | 1.38 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr21_-_23271127 | 1.36 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr25_-_35760878 | 1.34 |
ENSDART00000121786
|
hist1h4l
|
histone 1, H4, like |
| chr11_-_232269 | 1.29 |
ENSDART00000172818
|
si:ch1073-456m8.1
|
si:ch1073-456m8.1 |
| chr9_+_38975508 | 1.25 |
|
|
|
| chr14_+_35880163 | 1.24 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr7_+_26357875 | 1.21 |
ENSDART00000101044
|
hsbp1a
|
heat shock factor binding protein 1a |
| chr6_-_39315024 | 1.19 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr2_-_18157106 | 1.18 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr3_+_32360862 | 1.15 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr23_-_39956151 | 1.11 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr16_-_12621624 | 1.10 |
ENSDART00000155829
|
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr19_-_11113032 | 1.10 |
ENSDART00000027598
|
tpm3
|
tropomyosin 3 |
| chr12_-_20181519 | 1.07 |
ENSDART00000040909
|
rhbdf1b
|
rhomboid 5 homolog 1b (Drosophila) |
| chr5_-_39910235 | 1.07 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr11_+_3186396 | 1.04 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr7_+_25649559 | 1.03 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr6_-_22504772 | 1.02 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr23_+_2597825 | 0.99 |
ENSDART00000159039
|
ENSDARG00000089986
|
ENSDARG00000089986 |
| chr25_+_35796086 | 0.99 |
ENSDART00000112196
|
HIST2H4B
|
zgc:163040 |
| chr24_+_37518715 | 0.99 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr3_-_48865474 | 0.98 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr14_-_2682064 | 0.98 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr6_+_42821679 | 0.97 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr11_+_23522743 | 0.97 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr12_-_30662709 | 0.97 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr13_+_25298383 | 0.96 |
|
|
|
| chr4_+_1750689 | 0.95 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr10_+_45499009 | 0.94 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr16_+_28819826 | 0.94 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr3_+_14238188 | 0.92 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr5_+_71367929 | 0.92 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr3_-_39346621 | 0.91 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr17_-_20877606 | 0.90 |
ENSDART00000088106
|
ank3a
|
ankyrin 3a |
| chr25_+_35765309 | 0.88 |
ENSDART00000128547
|
hist1h4l
|
histone 1, H4, like |
| chr21_-_43020159 | 0.88 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr19_+_341775 | 0.88 |
ENSDART00000151013
|
ensaa
|
endosulfine alpha a |
| chr16_+_26989962 | 0.88 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr20_-_53560663 | 0.87 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr1_+_46768482 | 0.86 |
ENSDART00000015046
|
fstl1a
|
follistatin-like 1a |
| chr24_+_35676505 | 0.86 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr21_+_18295124 | 0.86 |
ENSDART00000167511
|
ENSDARG00000104101
|
ENSDARG00000104101 |
| chr12_+_689413 | 0.85 |
ENSDART00000174804
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr16_-_26803204 | 0.85 |
ENSDART00000162665
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr1_-_416138 | 0.85 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr25_-_35813232 | 0.85 |
ENSDART00000128564
|
hist1h4l
|
histone 1, H4, like |
| chr4_-_9172622 | 0.84 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr2_-_58111727 | 0.83 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr11_+_2786083 | 0.83 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr5_-_56984800 | 0.83 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr20_+_13998066 | 0.82 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr14_+_29428950 | 0.82 |
ENSDART00000105898
|
ENSDARG00000071567
|
ENSDARG00000071567 |
| chr14_-_36523075 | 0.81 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr15_+_30442890 | 0.80 |
ENSDART00000048847
|
nos2b
|
nitric oxide synthase 2b, inducible |
| chr22_-_35967525 | 0.80 |
ENSDART00000176971
|
CABZ01037120.1
|
ENSDARG00000106725 |
| chr10_+_10393377 | 0.80 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr16_+_17808623 | 0.79 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr4_+_7406743 | 0.79 |
ENSDART00000168327
|
cald1a
|
caldesmon 1a |
| chr7_+_6814828 | 0.79 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr22_+_1274718 | 0.78 |
ENSDART00000159296
|
si:ch73-138e16.4
|
si:ch73-138e16.4 |
| chr24_+_3296536 | 0.78 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr4_-_52067 | 0.78 |
ENSDART00000166186
|
RERG
|
RAS like estrogen regulated growth inhibitor |
| chr8_-_38357549 | 0.77 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr23_-_45637350 | 0.77 |
ENSDART00000149464
|
ENSDARG00000036897
|
ENSDARG00000036897 |
| chr20_-_47123598 | 0.75 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr3_-_60953138 | 0.75 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr23_-_624534 | 0.74 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr25_-_23428527 | 0.73 |
ENSDART00000062930
|
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr9_-_52847086 | 0.72 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr21_+_28921734 | 0.72 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr16_-_22218191 | 0.71 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr22_-_120677 | 0.71 |
|
|
|
| chr15_-_23407388 | 0.70 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr17_+_8166167 | 0.70 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr9_-_53220424 | 0.69 |
|
|
|
| chr6_+_56163589 | 0.69 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr20_-_54591757 | 0.69 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr5_+_22465722 | 0.69 |
ENSDART00000145477
ENSDART00000089992 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr24_+_3931981 | 0.68 |
ENSDART00000167043
|
pfkpa
|
phosphofructokinase, platelet a |
| chr23_+_39713307 | 0.68 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr25_-_34634503 | 0.68 |
ENSDART00000157370
|
CR762436.3
|
Histone H3.2 |
| chr5_-_71352034 | 0.68 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr2_-_1752776 | 0.67 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr8_+_20406797 | 0.66 |
ENSDART00000016422
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr9_-_30553051 | 0.66 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr24_+_2523708 | 0.65 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr13_+_48324283 | 0.65 |
ENSDART00000177067
|
CU929120.1
|
ENSDARG00000107321 |
| chr1_-_6976446 | 0.65 |
ENSDART00000085203
|
efnb2b
|
ephrin-B2b |
| chr3_+_16826320 | 0.62 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr18_+_16257606 | 0.62 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr15_-_16076874 | 0.62 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr16_+_53568814 | 0.62 |
ENSDART00000154189
|
rbm24b
|
RNA binding motif protein 24b |
| chr23_+_24557956 | 0.62 |
|
|
|
| chr5_+_50379968 | 0.61 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr3_-_5318206 | 0.61 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr19_+_5562107 | 0.61 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr14_-_45887823 | 0.60 |
ENSDART00000178932
ENSDART00000160657 |
cltb
|
clathrin, light chain B |
| chr16_+_51316772 | 0.59 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_+_1814994 | 0.59 |
|
|
|
| chr16_-_5205633 | 0.59 |
ENSDART00000082071
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr20_+_26408687 | 0.59 |
|
|
|
| chr12_+_13244482 | 0.59 |
ENSDART00000137757
|
irf9
|
interferon regulatory factor 9 |
| chr23_-_29630277 | 0.58 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr5_-_70939686 | 0.58 |
|
|
|
| chr7_-_69399273 | 0.58 |
ENSDART00000159823
ENSDART00000075178 ENSDART00000168942 ENSDART00000126739 |
tspan5a
|
tetraspanin 5a |
| chr7_+_13753344 | 0.57 |
|
|
|
| chr19_+_2013850 | 0.57 |
ENSDART00000048850
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr10_+_9259213 | 0.57 |
ENSDART00000064966
|
snx18b
|
sorting nexin 18b |
| chr14_+_9275853 | 0.56 |
ENSDART00000114563
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr3_+_12132746 | 0.56 |
|
|
|
| KN149874v1_+_3898 | 0.56 |
|
|
|
| chr2_-_1753058 | 0.56 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr18_+_512813 | 0.55 |
ENSDART00000162513
|
gpib
|
glucose-6-phosphate isomerase b |
| chr13_-_39821399 | 0.55 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr6_+_16279737 | 0.55 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr12_+_22448972 | 0.55 |
|
|
|
| chr3_+_22246697 | 0.54 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr16_+_40625705 | 0.54 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr3_+_59716594 | 0.54 |
ENSDART00000157351
ENSDART00000153928 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr2_+_26632673 | 0.54 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr19_+_43524098 | 0.54 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr14_+_891448 | 0.54 |
ENSDART00000021346
|
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr17_+_38314814 | 0.53 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr8_-_51859979 | 0.53 |
|
|
|
| chr1_-_22170553 | 0.53 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr3_-_7220340 | 0.53 |
|
|
|
| KN150266v1_+_74105 | 0.52 |
|
|
|
| chr16_+_5466342 | 0.52 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr17_+_12544451 | 0.51 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr6_-_2294751 | 0.51 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr12_-_48278273 | 0.50 |
|
|
|
| chr9_-_21420106 | 0.50 |
ENSDART00000102147
|
popdc2
|
popeye domain containing 2 |
| KN149874v1_+_3991 | 0.50 |
|
|
|
| chr10_+_5693933 | 0.50 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr5_-_25493792 | 0.50 |
ENSDART00000010199
|
fam219ab
|
family with sequence similarity 219, member Ab |
| chr15_+_46129807 | 0.50 |
ENSDART00000153936
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr15_-_21003820 | 0.50 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr19_+_14669633 | 0.49 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr3_+_14151127 | 0.49 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr25_-_34602827 | 0.49 |
ENSDART00000124121
|
hist1h4l
|
histone 1, H4, like |
| chr15_+_3296905 | 0.48 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr16_+_24766043 | 0.48 |
ENSDART00000114304
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr9_-_24602136 | 0.48 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr20_+_23392782 | 0.48 |
|
|
|
| chr11_-_23954234 | 0.47 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr13_-_37527812 | 0.47 |
ENSDART00000143806
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr8_+_22268070 | 0.47 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr22_-_62082 | 0.47 |
ENSDART00000166010
ENSDART00000125700 |
mrpl20
|
mitochondrial ribosomal protein L20 |
| chr11_+_6809190 | 0.46 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr24_-_14567403 | 0.46 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| KN150423v1_-_9765 | 0.46 |
|
|
|
| KN149965v1_-_10589 | 0.46 |
|
|
|
| chr1_-_40557436 | 0.46 |
ENSDART00000144424
|
rgs12b
|
regulator of G protein signaling 12b |
| chr3_-_30554400 | 0.46 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr9_-_30463616 | 0.46 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr20_-_28739099 | 0.46 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr5_+_15167637 | 0.45 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr16_-_6881218 | 0.45 |
ENSDART00000149070
|
mbpb
|
myelin basic protein b |
| chr23_+_10552781 | 0.45 |
|
|
|
| chr16_+_29623958 | 0.45 |
ENSDART00000088146
|
ensab
|
endosulfine alpha b |
| chr23_-_7889744 | 0.45 |
ENSDART00000164117
|
myt1b
|
myelin transcription factor 1b |
| chr19_+_48499602 | 0.44 |
|
|
|
| chr21_-_7277655 | 0.44 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr7_-_26137243 | 0.44 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr22_+_13952935 | 0.44 |
ENSDART00000080313
|
arl4ca
|
ADP-ribosylation factor-like 4Ca |
| chr8_+_14120313 | 0.44 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr13_+_25356184 | 0.44 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr5_-_20527105 | 0.42 |
ENSDART00000133461
|
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr21_+_7578326 | 0.42 |
ENSDART00000164883
|
CU655836.1
|
ENSDARG00000105204 |
| chr21_+_26684617 | 0.42 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr15_-_44957335 | 0.41 |
|
|
|
| chr3_-_5318289 | 0.41 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr7_-_10318692 | 0.41 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr16_+_12740993 | 0.40 |
ENSDART00000128497
|
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr6_-_39315608 | 0.40 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr5_+_69401770 | 0.40 |
|
|
|
| chr3_+_40173549 | 0.40 |
ENSDART00000083241
|
slc29a4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr1_-_46292847 | 0.40 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr23_+_23305483 | 0.39 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr8_-_53584884 | 0.39 |
|
|
|
| chr19_-_617298 | 0.39 |
ENSDART00000171387
|
cyp51
|
cytochrome P450, family 51 |
| chr21_+_13196717 | 0.39 |
ENSDART00000163767
|
adora2ab
|
adenosine A2a receptor b |
| chr3_+_40667131 | 0.39 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr23_-_36982827 | 0.39 |
ENSDART00000144333
|
cpsf3l
|
cleavage and polyadenylation specific factor 3-like |
| chr9_+_17341042 | 0.39 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr7_-_52223087 | 0.39 |
ENSDART00000129769
|
cgnl1
|
cingulin-like 1 |
| chr7_-_6322397 | 0.39 |
ENSDART00000172861
|
zgc:154164
|
zgc:154164 |
| chr4_-_72067910 | 0.39 |
ENSDART00000170308
|
BX855614.3
|
ENSDARG00000101193 |
| chr12_+_23641904 | 0.39 |
ENSDART00000152942
|
jcada
|
junctional cadherin 5 associated a |
| chr7_-_52574822 | 0.39 |
ENSDART00000172951
|
map1aa
|
microtubule-associated protein 1Aa |
| chr2_-_24947660 | 0.38 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr1_-_44239584 | 0.38 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.4 | 1.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.4 | 1.2 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.3 | 1.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.3 | 1.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.3 | 1.2 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.3 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.3 | 0.9 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.3 | 0.8 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.3 | 0.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.3 | 0.8 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.3 | 1.8 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.2 | 0.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 0.8 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.2 | 1.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 0.6 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 0.5 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.2 | 1.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.5 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.5 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 2.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.5 | GO:0071632 | optomotor response(GO:0071632) |
| 0.1 | 0.2 | GO:0050886 | endocrine process(GO:0050886) |
| 0.1 | 1.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.7 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.0 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.3 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 0.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.6 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.7 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.9 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.2 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.1 | 0.3 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) facial nerve morphogenesis(GO:0021610) |
| 0.1 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:0021572 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.1 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.2 | GO:0019320 | hexose catabolic process(GO:0019320) |
| 0.1 | 0.2 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.1 | 1.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 4.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.1 | 0.5 | GO:0035637 | multicellular organismal signaling(GO:0035637) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.7 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.9 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.3 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.1 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.3 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 1.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0060975 | cardioblast migration(GO:0003260) cardioblast migration to the midline involved in heart field formation(GO:0060975) |
| 0.0 | 0.1 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.1 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.6 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.8 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.0 | 0.6 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0072102 | glomerulus morphogenesis(GO:0072102) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.3 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.6 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.0 | GO:0010878 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.2 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 5.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 1.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 0.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 0.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 0.9 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 3.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.6 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 0.8 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 1.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.1 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.8 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0070548 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.9 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.8 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |