Project
DANIO-CODE
Navigation
Downloads
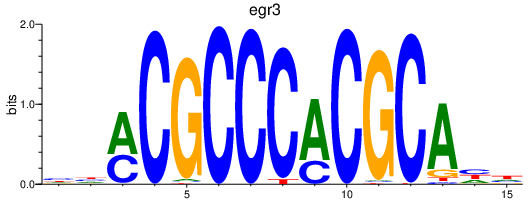
Results for egr3
Z-value: 0.51
Transcription factors associated with egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr3
|
ENSDARG00000089156 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr3 | dr10_dc_chr8_+_50738477_50738508 | -0.29 | 2.8e-01 | Click! |
Activity profile of egr3 motif
Sorted Z-values of egr3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of egr3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_47446494 | 1.50 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr7_+_45747395 | 1.42 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr13_+_35799602 | 1.37 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr7_+_45747622 | 1.33 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr19_-_48327666 | 1.28 |
|
|
|
| chr13_+_35799681 | 1.23 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr6_+_48188448 | 1.16 |
|
|
|
| chr3_-_5157662 | 1.14 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr22_+_37688839 | 1.09 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr6_+_48188313 | 1.03 |
|
|
|
| chr23_-_31719203 | 1.02 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_46403915 | 0.94 |
ENSDART00000154499
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| KN150034v1_+_1223 | 0.91 |
|
|
|
| chr9_-_55878791 | 0.83 |
|
|
|
| chr13_+_48475352 | 0.81 |
ENSDART00000165051
|
CU651669.1
|
ENSDARG00000101069 |
| chr15_-_14102102 | 0.81 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr12_-_37274632 | 0.78 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr13_-_36015558 | 0.77 |
ENSDART00000133072
|
med6
|
mediator complex subunit 6 |
| chr21_-_41508736 | 0.75 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr22_-_10562118 | 0.72 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| KN150034v1_+_1158 | 0.72 |
|
|
|
| chr6_+_48188520 | 0.70 |
|
|
|
| chr7_+_55217087 | 0.68 |
ENSDART00000149915
|
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr6_+_48188549 | 0.68 |
|
|
|
| chr22_+_37689055 | 0.65 |
|
|
|
| chr13_+_35799851 | 0.64 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr6_-_53391354 | 0.62 |
ENSDART00000155483
|
si:ch211-161c3.6
|
si:ch211-161c3.6 |
| chr3_-_45358502 | 0.60 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr18_+_41559512 | 0.60 |
ENSDART00000059135
|
bcl7bb
|
B-cell CLL/lymphoma 7B, b |
| chr4_-_5010148 | 0.56 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr15_+_46093261 | 0.56 |
|
|
|
| chr2_+_38959335 | 0.52 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr5_-_6054581 | 0.50 |
|
|
|
| chr12_-_2638179 | 0.44 |
|
|
|
| chr7_+_41060622 | 0.43 |
ENSDART00000160143
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr21_-_43402921 | 0.42 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr3_+_22853476 | 0.41 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr8_-_31374705 | 0.40 |
ENSDART00000162872
|
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr2_+_17042235 | 0.40 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr9_-_55878468 | 0.38 |
|
|
|
| chr25_+_1223427 | 0.38 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr15_+_14925529 | 0.38 |
ENSDART00000166250
|
CR936977.2
|
ENSDARG00000103638 |
| chr5_+_2472901 | 0.38 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr19_-_25842574 | 0.37 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr4_-_71945633 | 0.37 |
ENSDART00000170170
|
znf1015
|
zinc finger protein 1015 |
| chr10_-_8764361 | 0.36 |
|
|
|
| chr16_+_11027467 | 0.33 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr5_+_29231012 | 0.31 |
ENSDART00000159327
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr6_+_48188609 | 0.30 |
|
|
|
| chr13_-_36015609 | 0.30 |
ENSDART00000044115
|
med6
|
mediator complex subunit 6 |
| chr3_-_5318147 | 0.24 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr5_+_29231241 | 0.24 |
ENSDART00000131687
ENSDART00000035098 |
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr19_+_5562075 | 0.24 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr7_-_6228307 | 0.22 |
ENSDART00000173332
|
zgc:112234
|
zgc:112234 |
| chr12_-_37274547 | 0.21 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr22_+_37688988 | 0.20 |
|
|
|
| chr7_-_45747162 | 0.20 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr1_+_11608323 | 0.15 |
ENSDART00000164045
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr19_+_1298795 | 0.12 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr7_-_45747112 | 0.12 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr14_+_25866966 | 0.12 |
|
|
|
| chr7_+_41060426 | 0.11 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr22_+_37688695 | 0.09 |
ENSDART00000174664
ENSDART00000178200 |
FO704589.1
|
ENSDARG00000108910 |
| chr22_-_6287015 | 0.08 |
ENSDART00000148832
|
AL928685.4
|
ENSDARG00000095907 |
| chr16_+_11027394 | 0.07 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr15_+_9318502 | 0.07 |
ENSDART00000133588
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr3_-_11657881 | 0.06 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr11_-_2524380 | 0.06 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr7_-_6273677 | 0.05 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr21_+_6107404 | 0.03 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr18_+_41559549 | 0.02 |
ENSDART00000059135
|
bcl7bb
|
B-cell CLL/lymphoma 7B, b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.2 | 0.9 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.5 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.5 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.6 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 1.0 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 3.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.6 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 1.1 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.0 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.2 | 1.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |