Project
DANIO-CODE
Navigation
Downloads
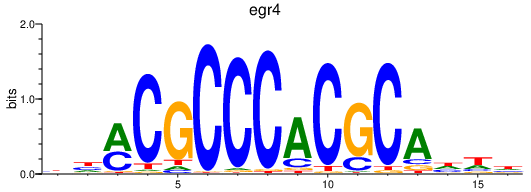
Results for egr4
Z-value: 1.26
Transcription factors associated with egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr4
|
ENSDARG00000077799 | early growth response 4 |
Activity profile of egr4 motif
Sorted Z-values of egr4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of egr4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_35060298 | 3.84 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr12_+_30389706 | 3.28 |
|
|
|
| chr16_+_27680365 | 3.15 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr13_+_35799681 | 3.11 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr14_+_14850200 | 2.92 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr16_-_47446494 | 2.85 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr6_+_48188448 | 2.78 |
|
|
|
| chr6_+_48188313 | 2.75 |
|
|
|
| chr14_-_35060332 | 2.61 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr13_+_35799602 | 2.61 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr17_+_46403915 | 2.49 |
ENSDART00000154499
|
si:dkey-206p8.1
|
si:dkey-206p8.1 |
| chr3_-_5157662 | 2.39 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| KN150034v1_+_1223 | 2.31 |
|
|
|
| KN150034v1_+_1158 | 2.19 |
|
|
|
| chr22_-_10562118 | 2.07 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr24_+_39285121 | 2.07 |
|
|
|
| chr8_-_49227286 | 2.05 |
|
|
|
| chr18_+_2124267 | 2.04 |
|
|
|
| chr8_-_49227232 | 1.92 |
|
|
|
| chr3_-_2041373 | 1.83 |
ENSDART00000146552
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr13_+_48475352 | 1.78 |
ENSDART00000165051
|
CU651669.1
|
ENSDARG00000101069 |
| chr6_+_48188520 | 1.73 |
|
|
|
| chr22_-_22315679 | 1.70 |
ENSDART00000033479
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr16_+_53632289 | 1.69 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr13_-_31267459 | 1.61 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr2_-_57502821 | 1.57 |
|
|
|
| chr2_+_38959335 | 1.48 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr6_+_48188549 | 1.47 |
|
|
|
| chr2_-_57502700 | 1.46 |
|
|
|
| chr13_-_31267133 | 1.41 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr19_-_48327666 | 1.38 |
|
|
|
| chr16_+_53632152 | 1.36 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr11_+_12662209 | 1.33 |
ENSDART00000054837
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr14_-_38549466 | 1.30 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr5_-_23192997 | 1.27 |
ENSDART00000167629
|
rnf128a
|
ring finger protein 128a |
| chr19_+_26807634 | 1.21 |
|
|
|
| chr22_-_9809500 | 1.20 |
|
|
|
| chr22_-_11024649 | 1.18 |
ENSDART00000105823
ENSDART00000159995 |
insrb
|
insulin receptor b |
| chr11_-_36425176 | 1.17 |
ENSDART00000156166
|
CR457444.1
|
ENSDARG00000096865 |
| chr3_+_29338315 | 1.15 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr3_+_22853476 | 1.11 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr2_+_17042235 | 1.08 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr12_-_7604946 | 1.08 |
ENSDART00000126712
|
ccdc6b
|
coiled-coil domain containing 6b |
| chr11_-_3240247 | 1.07 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr1_-_9789386 | 1.06 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr13_+_35799851 | 1.04 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr5_-_23192934 | 1.02 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr8_+_23144448 | 1.01 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr21_-_43402921 | 0.99 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr18_+_20493237 | 0.99 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr3_-_45358502 | 0.96 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr13_-_31267513 | 0.96 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr20_-_36510718 | 0.93 |
|
|
|
| chr11_+_12662507 | 0.92 |
ENSDART00000135761
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr20_-_10077762 | 0.89 |
ENSDART00000134439
|
znf770
|
zinc finger protein 770 |
| chr22_-_22315810 | 0.84 |
ENSDART00000033479
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr19_-_25842574 | 0.84 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr5_+_2472901 | 0.84 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr16_+_19223683 | 0.80 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr8_-_44617200 | 0.77 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr5_+_61279071 | 0.76 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr7_-_44623926 | 0.75 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr18_+_8943793 | 0.71 |
ENSDART00000144247
|
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr15_+_12127861 | 0.70 |
|
|
|
| chr7_+_41060622 | 0.67 |
ENSDART00000160143
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr3_-_5318147 | 0.66 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr18_+_2124217 | 0.66 |
|
|
|
| chr16_+_53632229 | 0.64 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr6_+_48188609 | 0.61 |
|
|
|
| chr19_+_5562075 | 0.58 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr12_-_7604882 | 0.56 |
ENSDART00000126219
|
ccdc6b
|
coiled-coil domain containing 6b |
| chr22_-_3165441 | 0.56 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr3_+_34690574 | 0.55 |
|
|
|
| chr13_+_11741597 | 0.50 |
|
|
|
| chr18_+_20493291 | 0.49 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr15_+_14925529 | 0.48 |
ENSDART00000166250
|
CR936977.2
|
ENSDARG00000103638 |
| chr5_+_61278798 | 0.45 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr8_-_49227143 | 0.42 |
|
|
|
| chr18_-_45620179 | 0.38 |
ENSDART00000163794
ENSDART00000146543 |
wt1b
|
wilms tumor 1b |
| chr22_+_10683985 | 0.35 |
ENSDART00000063207
ENSDART00000122349 |
hiat1b
|
hippocampus abundant transcript 1b |
| chr15_+_9318502 | 0.32 |
ENSDART00000133588
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr5_-_43219303 | 0.29 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr22_+_12773735 | 0.28 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr13_+_48191437 | 0.27 |
ENSDART00000053332
|
FOXN2 (1 of many)
|
forkhead box N2 |
| chr20_-_16366248 | 0.26 |
|
|
|
| chr16_-_40558536 | 0.25 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr7_+_41060426 | 0.23 |
ENSDART00000005127
|
garem
|
GRB2 associated, regulator of MAPK1 |
| chr5_+_61278989 | 0.23 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr10_+_19597171 | 0.22 |
ENSDART00000063810
ENSDART00000063806 |
atp6v1b2
|
ATPase, H+ transporting, lysosomal, V1 subunit B2 |
| chr17_+_26736073 | 0.19 |
ENSDART00000163886
|
larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr11_+_12662284 | 0.19 |
ENSDART00000054837
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr22_-_20695416 | 0.15 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr1_+_11608323 | 0.14 |
ENSDART00000164045
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr20_-_16366304 | 0.10 |
|
|
|
| chr21_+_6107404 | 0.09 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr3_-_5318012 | 0.08 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr8_+_53919318 | 0.08 |
|
|
|
| chr6_-_18019342 | 0.07 |
ENSDART00000154941
|
ten1
|
TEN1 CST complex subunit |
| chr8_-_53584884 | 0.04 |
|
|
|
| chr13_-_36483866 | 0.03 |
|
|
|
| chr21_+_6107190 | 0.01 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:1990379 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.9 | 6.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.6 | 2.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.6 | 3.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 2.5 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 0.9 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.2 | 1.0 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 2.4 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.1 | 1.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.3 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 1.1 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0090316 | positive regulation of intracellular protein transport(GO:0090316) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.3 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 6.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.1 | 3.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 2.4 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 2.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 2.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.7 | 4.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.4 | 2.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 3.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 6.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 1.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 2.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |