Project
DANIO-CODE
Navigation
Downloads
Results for ehf
Z-value: 1.41
Transcription factors associated with ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ehf
|
ENSDARG00000052115 | ets homologous factor |
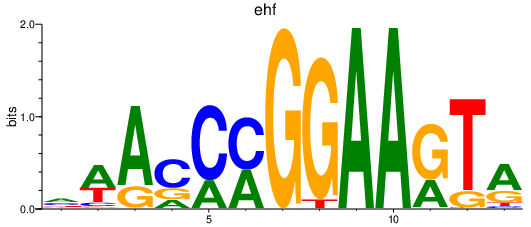
Activity profile of ehf motif
Sorted Z-values of ehf motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ehf
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_46426385 | 4.71 |
ENSDART00000141331
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr19_-_27093095 | 4.50 |
|
|
|
| chr10_-_17214368 | 3.96 |
|
|
|
| chr17_-_45021393 | 3.05 |
|
|
|
| chr24_-_9857510 | 2.82 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr1_-_21961282 | 2.75 |
ENSDART00000171830
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr25_+_4888647 | 2.74 |
ENSDART00000165933
ENSDART00000159537 |
hdac10
|
histone deacetylase 10 |
| KN150623v1_+_258 | 2.73 |
|
|
|
| chr1_-_7869105 | 2.59 |
ENSDART00000147763
|
zgc:112980
|
zgc:112980 |
| chr22_-_17627900 | 2.59 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr8_-_49227286 | 2.34 |
|
|
|
| chr8_-_49227232 | 2.31 |
|
|
|
| chr3_-_39117855 | 2.29 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| chr2_+_37224986 | 2.19 |
ENSDART00000138952
|
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr5_-_15971338 | 2.19 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr16_-_29755471 | 2.08 |
ENSDART00000133965
|
scnm1
|
sodium channel modifier 1 |
| chr25_-_3619590 | 2.07 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr4_-_14208573 | 2.06 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr22_-_17627831 | 2.06 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr1_+_18956542 | 1.97 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr24_+_31522539 | 1.96 |
ENSDART00000126380
|
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr1_+_18956648 | 1.94 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr14_-_6901209 | 1.93 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr1_-_40671573 | 1.92 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr20_-_23354440 | 1.90 |
ENSDART00000103365
|
ociad1
|
OCIA domain containing 1 |
| chr17_-_25313024 | 1.84 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr23_-_32267833 | 1.82 |
|
|
|
| chr23_+_25952724 | 1.82 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr16_-_25318413 | 1.81 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr17_-_9806363 | 1.81 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr3_+_27582277 | 1.80 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr9_+_29709931 | 1.76 |
ENSDART00000078904
ENSDART00000150164 |
fdx1
|
ferredoxin 1 |
| chr1_+_32825950 | 1.74 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr19_+_43468369 | 1.72 |
ENSDART00000165202
|
pum1
|
pumilio RNA-binding family member 1 |
| chr10_-_1933874 | 1.70 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr17_+_24791024 | 1.67 |
ENSDART00000130871
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr10_+_10354740 | 1.67 |
ENSDART00000142895
|
urm1
|
ubiquitin related modifier 1 |
| chr1_+_157840 | 1.66 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr24_-_9874069 | 1.66 |
ENSDART00000176344
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr15_+_38397715 | 1.64 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr13_-_9034919 | 1.63 |
ENSDART00000135373
|
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr7_-_48532462 | 1.63 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr8_+_8605240 | 1.61 |
ENSDART00000075624
|
usp11
|
ubiquitin specific peptidase 11 |
| chr23_+_25952958 | 1.60 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr23_+_25952913 | 1.59 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr5_+_24580551 | 1.59 |
ENSDART00000134526
|
paxx
|
zgc:193538 |
| chr11_+_2527564 | 1.59 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr6_-_3347207 | 1.58 |
ENSDART00000151416
|
si:ch1073-342h5.2
|
si:ch1073-342h5.2 |
| chr18_-_26815192 | 1.57 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr23_+_33792145 | 1.56 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr5_+_19429620 | 1.55 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr5_+_9579430 | 1.55 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr17_+_24790812 | 1.53 |
ENSDART00000082251
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr13_-_37340209 | 1.52 |
|
|
|
| chr10_-_180603 | 1.51 |
|
|
|
| chr17_+_25313170 | 1.50 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr7_-_26332194 | 1.49 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr7_+_69290790 | 1.49 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr3_-_32457708 | 1.49 |
|
|
|
| chr7_-_6334676 | 1.47 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr20_+_34084660 | 1.47 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr24_-_37450413 | 1.47 |
ENSDART00000056303
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr23_+_17596504 | 1.45 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr16_-_52847564 | 1.44 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr10_-_1933761 | 1.44 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr25_-_21409895 | 1.43 |
ENSDART00000010706
|
immp2l
|
inner mitochondrial membrane peptidase subunit 2 |
| chr5_+_40235387 | 1.43 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr7_+_73598715 | 1.43 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr14_-_20594454 | 1.42 |
ENSDART00000132499
|
ids
|
iduronate 2-sulfatase |
| chr19_+_43928043 | 1.41 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr15_-_29393378 | 1.41 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr3_+_17901295 | 1.41 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr20_-_18482817 | 1.40 |
ENSDART00000159636
|
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr1_-_52833379 | 1.40 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr7_-_6618142 | 1.40 |
ENSDART00000002949
|
ENSDARG00000045199
|
ENSDARG00000045199 |
| chr24_-_23570846 | 1.38 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr8_+_41004169 | 1.37 |
|
|
|
| chr14_-_20940726 | 1.37 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr8_+_41003546 | 1.37 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr24_+_37450437 | 1.36 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr6_-_40715613 | 1.35 |
ENSDART00000153702
|
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr16_+_43464824 | 1.34 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr11_+_13118866 | 1.33 |
ENSDART00000123257
|
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr11_-_25615491 | 1.33 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr18_-_49291686 | 1.32 |
ENSDART00000174038
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr14_+_35084220 | 1.31 |
ENSDART00000171809
|
sytl4
|
synaptotagmin-like 4 |
| chr15_+_38397897 | 1.31 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr20_+_29685379 | 1.31 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr25_-_25480657 | 1.30 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr20_+_24047842 | 1.28 |
ENSDART00000128616
ENSDART00000144195 |
casp8ap2
|
caspase 8 associated protein 2 |
| chr7_-_57876365 | 1.27 |
ENSDART00000073635
|
nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr1_+_46474561 | 1.27 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr2_-_41888389 | 1.25 |
|
|
|
| chr13_+_25418877 | 1.25 |
ENSDART00000169199
|
sec23ip
|
SEC23 interacting protein |
| chr24_-_37438115 | 1.24 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr21_-_43654780 | 1.24 |
ENSDART00000142733
|
si:dkey-229d11.5
|
si:dkey-229d11.5 |
| chr5_-_56953587 | 1.23 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr20_-_3220475 | 1.23 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr8_-_47339228 | 1.23 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr17_-_35331055 | 1.22 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr17_-_31195299 | 1.21 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr3_-_36298676 | 1.21 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr5_+_51186708 | 1.20 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr19_+_47918720 | 1.20 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr4_-_12979905 | 1.19 |
ENSDART00000150674
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr13_-_25277861 | 1.19 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr7_-_13416823 | 1.19 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| chr3_-_58400454 | 1.18 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr20_+_31036284 | 1.18 |
ENSDART00000153344
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr24_-_32607801 | 1.17 |
ENSDART00000143781
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr13_-_22831037 | 1.17 |
ENSDART00000057641
|
tspan15
|
tetraspanin 15 |
| chr4_+_1873500 | 1.17 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr23_+_17596460 | 1.15 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr17_-_31195376 | 1.14 |
ENSDART00000086511
|
rpusd2
|
RNA pseudouridylate synthase domain containing 2 |
| chr13_-_22831005 | 1.14 |
ENSDART00000143112
|
tspan15
|
tetraspanin 15 |
| chr8_-_47339136 | 1.14 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr23_+_28464194 | 1.13 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr6_+_3556296 | 1.13 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr24_+_16575167 | 1.13 |
|
|
|
| chr7_+_40878834 | 1.13 |
|
|
|
| chr3_-_5157662 | 1.12 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr20_+_34598242 | 1.11 |
ENSDART00000143765
|
gorab
|
golgin, rab6-interacting |
| chr24_-_1014256 | 1.11 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr24_-_1014491 | 1.11 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr6_+_52302917 | 1.10 |
|
|
|
| chr5_-_34856057 | 1.10 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr5_+_19363942 | 1.10 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr11_-_7786415 | 1.09 |
ENSDART00000154569
|
CR450813.1
|
ENSDARG00000097035 |
| chr13_-_24265471 | 1.09 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr14_-_5100304 | 1.09 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr8_-_49227143 | 1.07 |
|
|
|
| chr21_+_20350218 | 1.07 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr24_-_1014640 | 1.06 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr6_+_49772891 | 1.06 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr5_+_19429500 | 1.05 |
ENSDART00000168868
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr15_-_35020312 | 1.05 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr13_-_44694139 | 1.04 |
ENSDART00000137275
|
si:dkeyp-2e4.6
|
si:dkeyp-2e4.6 |
| chr3_-_34208507 | 1.04 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr17_-_9806413 | 1.03 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr15_+_21776031 | 1.03 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr17_-_20198477 | 1.02 |
ENSDART00000063523
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr14_-_27040866 | 1.02 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr3_-_26935578 | 1.02 |
ENSDART00000156311
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr25_-_24150237 | 1.02 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr10_-_180681 | 1.01 |
|
|
|
| chr6_-_10492276 | 1.00 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr20_+_46668743 | 0.99 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr5_+_71100066 | 0.99 |
ENSDART00000115182
|
nup214
|
nucleoporin 214 |
| chr19_-_15324823 | 0.99 |
ENSDART00000169883
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr5_-_56810425 | 0.98 |
ENSDART00000147875
ENSDART00000142776 |
ENSDARG00000095415
|
ENSDARG00000095415 |
| chr6_-_44164161 | 0.98 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr5_+_51186666 | 0.97 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr13_+_34563757 | 0.97 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr25_-_4407821 | 0.97 |
|
|
|
| chr9_-_32994156 | 0.97 |
ENSDART00000133382
|
zgc:112056
|
zgc:112056 |
| chr5_-_9443859 | 0.97 |
|
|
|
| chr3_+_27582359 | 0.97 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr7_-_6618225 | 0.96 |
ENSDART00000113658
|
ENSDARG00000045199
|
ENSDARG00000045199 |
| chr10_+_30640 | 0.94 |
ENSDART00000040240
|
tiprl
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr19_+_30835769 | 0.94 |
ENSDART00000023074
|
mrps18b
|
mitochondrial ribosomal protein S18B |
| chr21_+_6008781 | 0.94 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr13_+_11307037 | 0.94 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_+_20901498 | 0.93 |
|
|
|
| chr16_+_29756032 | 0.92 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr3_+_19515578 | 0.92 |
ENSDART00000007857
|
mettl2a
|
methyltransferase like 2A |
| chr15_+_46093261 | 0.92 |
|
|
|
| chr10_-_36149719 | 0.92 |
|
|
|
| chr21_-_43671213 | 0.90 |
ENSDART00000139008
|
si:dkey-229d11.3
|
si:dkey-229d11.3 |
| chr5_-_56953716 | 0.90 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr14_-_21779536 | 0.90 |
|
|
|
| chr16_-_25318334 | 0.90 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr6_+_48188313 | 0.89 |
|
|
|
| chr1_-_40671522 | 0.89 |
ENSDART00000074777
ENSDART00000158114 |
htt
|
huntingtin |
| chr10_-_7677571 | 0.89 |
ENSDART00000166234
|
ubxn8
|
UBX domain protein 8 |
| chr7_+_38257950 | 0.88 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr11_+_7173167 | 0.88 |
ENSDART00000125619
|
thop1
|
thimet oligopeptidase 1 |
| chr16_-_13723778 | 0.88 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr7_+_17460777 | 0.88 |
|
|
|
| chr15_-_35020250 | 0.88 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr23_+_28464298 | 0.87 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr7_-_5253629 | 0.87 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr19_-_12158958 | 0.87 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr13_-_43503453 | 0.87 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr13_-_42598533 | 0.87 |
ENSDART00000160472
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr2_-_32278726 | 0.87 |
ENSDART00000159843
ENSDART00000056621 ENSDART00000039717 ENSDART00000145704 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr16_-_25317921 | 0.86 |
|
|
|
| chr6_+_48188448 | 0.86 |
|
|
|
| chr21_+_19040595 | 0.86 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr6_+_23218068 | 0.86 |
ENSDART00000164960
|
stx8
|
syntaxin 8 |
| chr19_+_1148544 | 0.85 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr7_-_66470118 | 0.85 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr8_+_50162137 | 0.85 |
ENSDART00000056074
|
entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr5_-_12242927 | 0.84 |
ENSDART00000137705
|
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr23_+_25953162 | 0.84 |
ENSDART00000138731
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr3_-_58400345 | 0.84 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr19_+_37548595 | 0.84 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr14_-_38606412 | 0.84 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr23_+_16880876 | 0.84 |
ENSDART00000141966
|
zgc:114081
|
zgc:114081 |
| chr16_-_25318260 | 0.83 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr8_+_39726564 | 0.83 |
ENSDART00000145112
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 1.0 | 3.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.0 | 2.9 | GO:0021990 | neural plate formation(GO:0021990) brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) anterior neural plate formation(GO:0090017) |
| 0.8 | 5.9 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.8 | 2.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.6 | 0.6 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.5 | 4.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.5 | 1.5 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.5 | 1.4 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.5 | 1.8 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.4 | 2.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.4 | 1.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.4 | 1.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 1.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 1.8 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.3 | 2.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 1.4 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.3 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.0 | GO:0043368 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) type 2 immune response(GO:0042092) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T cell selection(GO:0045058) T-helper 2 cell differentiation(GO:0045064) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.3 | 1.2 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.3 | 0.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.3 | 0.8 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.3 | 0.8 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.3 | 0.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 5.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 0.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.8 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.2 | 0.6 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 0.4 | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043370) regulation of alpha-beta T cell differentiation(GO:0046637) regulation of CD4-positive, alpha-beta T cell activation(GO:2000514) |
| 0.2 | 3.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 2.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 1.4 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.2 | 2.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 1.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 2.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.2 | 0.8 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 0.9 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 2.8 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.7 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.5 | GO:1903059 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.1 | 0.6 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.1 | 0.5 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 1.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 0.5 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.8 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.1 | 0.6 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.5 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.1 | 4.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 2.2 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 1.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 3.3 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.5 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.4 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.6 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 2.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 1.1 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.1 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.3 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 0.6 | GO:0045880 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.3 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 5.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.7 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 1.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 4.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.6 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.2 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0006544 | glycine metabolic process(GO:0006544) glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.6 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.2 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 5.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 1.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.6 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 3.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0071236 | response to antibiotic(GO:0046677) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.7 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 1.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.2 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 2.5 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 1.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.9 | GO:0019953 | sexual reproduction(GO:0019953) |
| 0.0 | 0.1 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0015804 | neutral amino acid transport(GO:0015804) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.3 | GO:0010469 | regulation of receptor activity(GO:0010469) |
| 0.0 | 0.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 1.1 | GO:0007219 | Notch signaling pathway(GO:0007219) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 4.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.3 | 2.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 1.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 1.6 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 2.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.6 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 4.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 4.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 3.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.4 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.5 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.7 | 2.0 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.6 | 2.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 2.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 3.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.5 | 1.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.5 | 1.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.5 | 1.8 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.4 | 4.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 1.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 3.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.4 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.3 | 1.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 0.6 | GO:0001016 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.3 | 3.1 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.3 | 1.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 0.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.9 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 1.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.2 | 1.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 0.9 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 1.9 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 0.6 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 0.8 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.6 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 1.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.9 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 4.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 2.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.3 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.1 | 2.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.4 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.2 | GO:0043878 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 0.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 3.1 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin-like protein binding(GO:0032182) ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 6.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 1.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 2.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 3.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 4.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 3.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.9 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 2.7 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |